Nasal and blood transcriptomic pathways underpinning the clinical response to grass pollen immunotherapy
- PMID: 37460024
- PMCID: PMC10788383
- DOI: 10.1016/j.jaci.2023.06.025
Nasal and blood transcriptomic pathways underpinning the clinical response to grass pollen immunotherapy
Abstract
Background: Allergen immunotherapy (AIT) is a well-established disease-modifying therapy for allergic rhinitis, yet the fundamental mechanisms underlying its clinical effect remain inadequately understood. Gauging Response in Allergic Rhinitis to Sublingual and Subcutaneous Immunotherapy was a randomized, double-blind, placebo-controlled trial of individuals allergic to timothy grass who received 2 years of placebo (n = 30), subcutaneous immunotherapy (SCIT) (n = 27), or sublingual immunotherapy (SLIT) (n = 27) and were then followed for 1 additional year.
Objective: We used yearly biospecimens from the Gauging Response in Allergic Rhinitis to Sublingual and Subcutaneous Immunotherapy study to identify molecular mechanisms of response.
Methods: We used longitudinal transcriptomic profiling of nasal brush and PBMC samples after allergen provocation to uncover airway and systemic expression pathways mediating responsiveness to AIT.
Trial registration: ClinicalTrials.gov Identifier: NCT01335139, EudraCT Number: 2010-023536-16.
Results: SCIT and SLIT demonstrated similar changes in gene module expression over time. In nasal samples, alterations included downregulation of pathways of mucus hypersecretion, leukocyte migration/activation, and endoplasmic reticulum stress (log2 fold changes -0.133 to -0.640, false discovery rates [FDRs] <0.05). We observed upregulation of modules related to epithelial development, junction formation, and lipid metabolism (log2 fold changes 0.104 to 0.393, FDRs <0.05). In PBMCs, modules related to cellular stress response and type 2 cytokine signaling were reduced by immunotherapy (log2 fold changes -0.611 to -0.828, FDRs <0.05). Expression of these modules was also significantly associated with both Total Nasal Symptom Score and peak nasal inspiratory flow, indicating important links between treatment, module expression, and allergen response.
Conclusions: Our results identify specific molecular responses of the nasal airway impacting barrier function, leukocyte migration activation, and mucus secretion that are affected by both SCIT and SLIT, offering potential targets to guide novel strategies for AIT.
Keywords: Allergen immunotherapy; RNA sequencing; allergic rhinitis; subcutaneous immunotherapy; sublingual immunotherapy.
Copyright © 2023 American Academy of Allergy, Asthma & Immunology. All rights reserved.
Conflict of interest statement
Disclaimer: Dr Togias’ co-authorship of this publication does not constitute an endorsement by the US National Institute of Allergy and Infectious Diseases, the National Institutes of Health or any other agency of the United States government.
GWS reports lecture fees from ALK-Abello and Mylan; sponsorship to attend a virtual conference (EAACI 2021) from GSK. MHS reports research grants Medical Research Council, Allergy Therapeutics, LETI Laboratorios, Revolo Biotherapeutics and lecture fees from Allergy Therapeutics and Leti Laboratorios. SRD has received research Grants from personal fees for consultancies from Revelo, ANGANY Inc., ALK and lecture fees from Abbott Laboratories, ALK, Allergopharma, Pneumo Update GmbH and Stallergenes. RMS, DL, NDJ, MS, SP, TQ, SS, ES, AOE, MP, MRJ, RVP, report no competing interests.
Figures


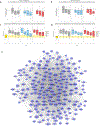




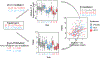
References
-
- Guerra S, Sherrill DL, Martinez FD, Barbee RA. Rhinitis as an independent risk factor for adult-onset asthma. Journal of Allergy and Clinical Immunology 2002; 109:419–25. - PubMed
-
- Shaaban R, Zureik M, Soussan D, Neukirch C, Heinrich J, Sunyer J, et al. Rhinitis and onset of asthma: a longitudinal population-based study. The Lancet 2008; 372:1049–57. - PubMed
-
- Larsen JN, Broge L, Jacobi H. Allergy immunotherapy: the future of allergy treatment. Drug Discov Today 2016; 21:26–37. - PubMed

