Single-cell transcriptomic profiles in the pathophysiology within the microenvironment of early diabetic kidney disease
- PMID: 37460555
- PMCID: PMC10352247
- DOI: 10.1038/s41419-023-05947-1
Single-cell transcriptomic profiles in the pathophysiology within the microenvironment of early diabetic kidney disease
Abstract
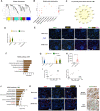
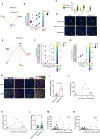
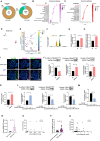
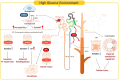
Diabetic kidney disease (DKD) is the leading cause of end-stage kidney disease, resulting in a huge socio-economic impact. Kidney is a highly complex organ and the pathogenesis underlying kidney organization involves complex cell-to-cell interaction within the heterogeneous kidney milieu. Advanced single-cell RNA sequencing (scRNA-seq) could reveal the complex architecture and interaction with the microenvironment in early DKD. We used scRNA-seq to investigate early changes in the kidney of db/m mice and db/db mice at the 14th week. Uniform Manifold Approximation and Projection were applied to classify cells into different clusters at a proper resolution. Weighted gene co-expression network analysis was used to identify the key molecules specifically expressed in kidney tubules. Information of cell-cell communication within the kidney was obtained using receptor-ligand pairing resources. In vitro model, human subjects, and co-detection by indexing staining were used to identify the pathophysiologic role of the hub genes in DKD. Among four distinct subsets of the proximal tubule (PT), lower percentages of proliferative PT and PT containing AQP4 expression (PTAQP4+) in db/db mice induced impaired cell repair activity and dysfunction of renin-angiotensin system modulation in early DKD. We found that ferroptosis was involved in DKD progression, and ceruloplasmin acted as a central regulator of the induction of ferroptosis in PTAQP4+. In addition, lower percentages of thick ascending limbs and collecting ducts with impaired metabolism function were also critical pathogenic features in the kidney of db/db mice. Secreted phosphoprotein 1 (SPP1) mediated pathogenic cross-talk in the tubular microenvironment, as validated by a correlation between urinary SPP1/Cr level and tubular injury. Finally, mesangial cell-derived semaphorin 3C (SEMA3C) further promoted endothelium-mesenchymal transition in glomerular endothelial cells through NRP1 and NRP2, and urinary SEMA3C/Cr level was positively correlated with glomerular injury. These data identified the hub genes involved in pathophysiologic changes within the microenvironment of early DKD.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

