Re-evaluation and re-analysis of 152 research exomes five years after the initial report reveals clinically relevant changes in 18
- PMID: 37460657
- PMCID: PMC10545662
- DOI: 10.1038/s41431-023-01425-6
Re-evaluation and re-analysis of 152 research exomes five years after the initial report reveals clinically relevant changes in 18
Abstract
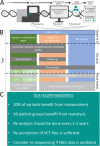
Iterative re-analysis of NGS results is not well investigated for published research cohorts of rare diseases. We revisited a cohort of 152 consanguineous families with developmental disorders (NDD) reported five years ago. We re-evaluated all reported variants according to diagnostic classification guidelines or our candidate gene scoring system (AutoCaSc) and systematically scored the validity of gene-disease associations (GDA). Sequencing data was re-processed using an up-to-date pipeline for case-level re-analysis. In 28/152 (18%) families, we identified a clinically relevant change. Ten previously reported (likely) pathogenic variants were re-classified as VUS/benign. In one case, the GDA (TSEN15) validity was judged as limited, and in five cases GDAs are meanwhile established. We identified 12 new disease causing variants. Two previously reported variants were missed by our updated pipeline due to alignment or reference issues. Our results support the need to re-evaluate screening studies, not only the negative cases but including supposedly solved ones. This also applies in a diagnostic setting. We highlight that the complexity of computational re-analysis for old data should be weighed against the decreasing re-testing costs. Since extensive re-analysis per case is beyond the resources of most institutions, we recommend a screening procedure that would quickly identify the majority (83%) of new variants.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
Unveiling the hidden: revisiting the potential of old genetic data for rare disease research.Eur J Hum Genet. 2023 Oct;31(10):1093-1094. doi: 10.1038/s41431-023-01435-4. Epub 2023 Jul 20. Eur J Hum Genet. 2023. PMID: 37474787 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

