AgAnimalGenomes: browsers for viewing and manually annotating farm animal genomes
- PMID: 37460664
- PMCID: PMC10382368
- DOI: 10.1007/s00335-023-10008-1
AgAnimalGenomes: browsers for viewing and manually annotating farm animal genomes
Abstract
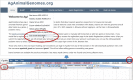
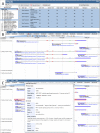
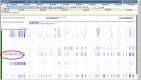
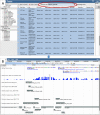
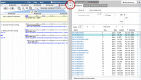
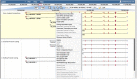
Current genome sequencing technologies have made it possible to generate highly contiguous genome assemblies for non-model animal species. Despite advances in genome assembly methods, there is still room for improvement in the delineation of specific gene features in the genomes. Here we present genome visualization and annotation tools to support seven livestock species (bovine, chicken, goat, horse, pig, sheep, and water buffalo), available in a new resource called AgAnimalGenomes. In addition to supporting the manual refinement of gene models, these browsers provide visualization tracks for hundreds of RNAseq experiments, as well as data generated by the Functional Annotation of Animal Genomes (FAANG) Consortium. For species with predicted gene sets from both Ensembl and RefSeq, the browsers provide special tracks showing the thousands of protein-coding genes that disagree across the two gene sources, serving as a valuable resource to alert researchers to gene model issues that may affect data interpretation. We describe the data and search methods available in the new genome browsers and how to use the provided tools to edit and create new gene models.
© 2023. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andersson L, Archibald AL, Bottema CD, Brauning R, Burgess SC, Burt DW, Casas E, Cheng HH, Clarke L, Couldrey C, Dalrymple BP, Elsik CG, Foissac S, Giuffra E, Groenen MA, Hayes BJ, Huang LS, Khatib H, Kijas JW, Kim H, Lunney JK, McCarthy FM, McEwan JC, Moore S, Nanduri B, Notredame C, Palti Y, Plastow GS, Reecy JM, Rohrer GA, Sarropoulou E, Schmidt CJ, Silverstein J, Tellam RL, Tixier-Boichard M, Tosser-Klopp G, Tuggle CK, Vilkki J, White SN, Zhao S, Zhou H, Consortium F. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 2015;16:57. - PMC - PubMed
-
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, Burton JN, Huson HJ, Nystrom JC, Kelley CM, Hutchison JL, Zhou Y, Sun J, Crisà A, Ponce de León FA, Schwartz JC, Hammond JA, Waldbieser GC, Schroeder SG, Liu GE, Dunham MJ, Shendure J, Sonstegard TS, Phillippy AM, Van Tassell CP, Smith TP. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49:643–650. - PMC - PubMed
-
- Bush SJ, McCulloch MEB, Muriuki C, Salavati M, Davis GM, Farquhar IL, Lisowski ZM, Archibald AL, Hume DA, Clark EL. Comprehensive transcriptional profiling of the gastrointestinal tract of ruminants from birth to adulthood reveals strong developmental stage specific gene expression. G3 (bethesda) 2019;9:359–373. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
