Toxicology knowledge graph for structural birth defects
- PMID: 37460679
- PMCID: PMC10352311
- DOI: 10.1038/s43856-023-00329-2
Toxicology knowledge graph for structural birth defects
Abstract
Background: Birth defects are functional and structural abnormalities that impact about 1 in 33 births in the United States. They have been attributed to genetic and other factors such as drugs, cosmetics, food, and environmental pollutants during pregnancy, but for most birth defects there are no known causes.
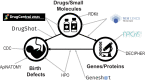
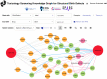
Methods: To further characterize associations between small molecule compounds and their potential to induce specific birth abnormalities, we gathered knowledge from multiple sources to construct a reproductive toxicity Knowledge Graph (ReproTox-KG) with a focus on associations between birth defects, drugs, and genes. Specifically, we gathered data from drug/birth-defect associations from co-mentions in published abstracts, gene/birth-defect associations from genetic studies, drug- and preclinical-compound-induced gene expression changes in cell lines, known drug targets, genetic burden scores for human genes, and placental crossing scores for small molecules.
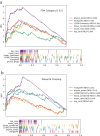
Results: Using ReproTox-KG and semi-supervised learning (SSL), we scored >30,000 preclinical small molecules for their potential to cross the placenta and induce birth defects, and identified >500 birth-defect/gene/drug cliques that can be used to explain molecular mechanisms for drug-induced birth defects. The ReproTox-KG can be accessed via a web-based user interface available at https://maayanlab.cloud/reprotox-kg . This site enables users to explore the associations between birth defects, approved and preclinical drugs, and all human genes.
Conclusions: ReproTox-KG provides a resource for exploring knowledge about the molecular mechanisms of birth defects with the potential of predicting the likelihood of genes and preclinical small molecules to induce birth defects.
Plain language summary
While birth defects are common, for most birth defects there are no known causes. During pregnancy, developing babies are exposed to drugs, cosmetics, food, and environmental pollutants that may cause birth defects. However, exactly how these environmental factors are involved in producing birth defects is difficult to discern. Also, birth defects can be a consequence of the genes inherited from the parents. We combined general data about human genes and drugs with specific data previously implicating genes and drugs in inducing birth defects to create a knowledge graph representation that connects genes, drugs, and birth defects. This knowledge graph can be used to explore new links that may explain why birth defects occur, particularly those that result from a combination of inherited and environmental influences.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Update on Overall Prevalence of Major Birth Defects. CDC MMWR 57, 1–5 (2008). - PubMed
-
- Principles For Evaluating Health Risks To Reproduction Associated With Exposure To Chemicals. (2001) https://inchem.org/documents/ehc/ehc/ehc225.htm.

