Interrogating colorectal cancer metastasis to liver: a search for clinically viable compounds and mechanistic insights in colorectal cancer Patient Derived Organoids
- PMID: 37460938
- PMCID: PMC10351152
- DOI: 10.1186/s13046-023-02754-6
Interrogating colorectal cancer metastasis to liver: a search for clinically viable compounds and mechanistic insights in colorectal cancer Patient Derived Organoids
Abstract
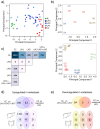
Background: Approximately 20-50% of patients presenting with localized colorectal cancer progress to stage IV metastatic disease (mCRC) following initial treatment and this is a major prognostic determinant. Here, we have interrogated a heterogeneous set of primary colorectal cancer (CRC), liver CRC metastases and adjacent liver tissue to identify molecular determinants of the colon to liver spreading. Screening Food and Drug Administration (FDA) approved drugs for their ability to interfere with an identified colon to liver metastasis signature may help filling an unmet therapeutic need.
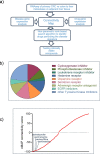
Methods: RNA sequencing of primary colorectal cancer specimens vs adjacent liver tissue vs synchronous and asynchronous liver metastases. Pathways enrichment analyses. The Library of Integrated Network-based Cellular Signatures (LINCS)-based and Connectivity Map (CMAP)-mediated identification of FDA-approved compounds capable to interfere with a 22 gene signature from primary CRC and liver metastases. Testing the identified compounds on CRC-Patient Derived Organoid (PDO) cultures. Microscopy and Fluorescence Activated Cell Sorting (FACS) based analysis of the treated PDOs.
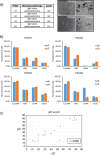
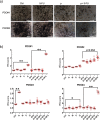
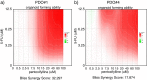
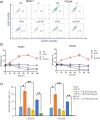
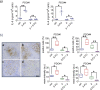
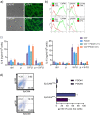
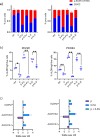
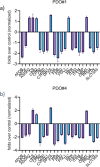
Results: We have found that liver metastases acquire features of the adjacent liver tissue while partially losing those of the primary tumors they derived from. We have identified a 22-gene signature differentially expressed among primary tumors and metastases and validated in public databases. A pharmacogenomic screening for FDA-approved compounds capable of interfering with this signature has been performed. We have validated some of the identified representative compounds in CRC-Patient Derived Organoid cultures (PDOs) and found that pentoxyfilline and, to a minor extent, dexketoprofen and desloratadine, can variably interfere with number, size and viability of the CRC -PDOs in a patient-specific way. We explored the pentoxifylline mechanism of action and found that pentoxifylline treatment attenuated the 5-FU elicited increase of ALDHhigh cells by attenuating the IL-6 mediated STAT3 (tyr705) phosphorylation.
Conclusions: Pentoxifylline synergizes with 5-Fluorouracil (5-FU) in attenuating organoid formation. It does so by interfering with an IL-6-STAT3 axis leading to the emergence of chemoresistant ALDHhigh cell subpopulations in 5-FU treated PDOs. A larger cohort of CRC-PDOs will be required to validate and expand on the findings of this proof-of-concept study.
Keywords: 5-FU; CMAP; CRC; Chemoresistance; Desloratadine; Dexketoprofen; IL-6; Liver metastases; Organoids; Pentoxifylline; STAT3.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

