A high fidelity approach to assembling the complex Borrelia genome
- PMID: 37460975
- PMCID: PMC10353223
- DOI: 10.1186/s12864-023-09500-4
A high fidelity approach to assembling the complex Borrelia genome
Abstract
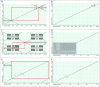
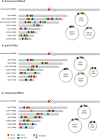
Background: Bacteria of the Borrelia burgdorferi sensu lato (s.l.) complex can cause Lyme borreliosis. Different B. burgdorferi s.l. genospecies vary in their host and vector associations and human pathogenicity but the genetic basis for these adaptations is unresolved and requires completed and reliable genomes for comparative analyses. The de novo assembly of a complete Borrelia genome is challenging due to the high levels of complexity, represented by a high number of circular and linear plasmids that are dynamic, showing mosaic structure and sequence homology. Previous work demonstrated that even advanced approaches, such as a combination of short-read and long-read data, might lead to incomplete plasmid reconstruction. Here, using recently developed high-fidelity (HiFi) PacBio sequencing, we explored strategies to obtain gap-free, complete and high quality Borrelia genome assemblies. Optimizing genome assembly, quality control and refinement steps, we critically appraised existing techniques to create a workflow that lead to improved genome reconstruction.
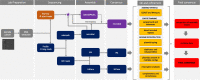
Results: Despite the latest available technologies, stand-alone sequencing and assembly methods are insufficient for the generation of complete and high quality Borrelia genome assemblies. We developed a workflow pipeline for the de novo genome assembly for Borrelia using several types of sequence data and incorporating multiple assemblers to recover the complete genome including both circular and linear plasmid sequences.
Conclusion: Our study demonstrates that, with HiFi data and an ensemble reconstruction pipeline with refinement steps, chromosomal and plasmid sequences can be fully resolved, even for complex genomes such as Borrelia. The presented pipeline may be of interest for the assembly of further complex microbial genomes.
Keywords: Borrelia burgdorferi; De novo assembly; Genome reconstruction pipeline; Genomics; HiFi sequencing; Plasmids.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Margos G, Henningsson AJ, Hepner S, Markowicz M, Sing A, Fingerle V. Borrelia ecology, evolution, and human disease: A mosaic of life. In: Sing A, editor. Zoonoses: Infections Affecting Humans and Animals. Cham: Springer International Publishing; 2022. pp. 1–66.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

