This is a preprint.
Microglia-derived TGF-β1 ligand maintains microglia homeostasis via autocrine mechanism and is critical for normal cognitive function in adult mouse brain
- PMID: 37461569
- PMCID: PMC10349967
- DOI: 10.1101/2023.07.05.547814
Microglia-derived TGF-β1 ligand maintains microglia homeostasis via autocrine mechanism and is critical for normal cognitive function in adult mouse brain
Update in
-
Adult microglial TGFβ1 is required for microglia homeostasis via an autocrine mechanism to maintain cognitive function in mice.Nat Commun. 2024 Jun 21;15(1):5306. doi: 10.1038/s41467-024-49596-0. Nat Commun. 2024. PMID: 38906887 Free PMC article.
Abstract
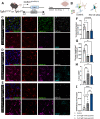
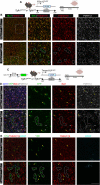
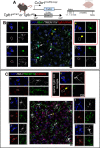
While TGF-β signaling is essential for microglial function, the cellular source of TGF-β ligand and its spatial regulation remains unclear in the adult CNS. Our data support that microglia, not astrocytes or neurons, are the primary producers of TGF-β1 ligands needed for microglial homeostasis. Microglia (MG)-Tgfb1 inducible knockout (iKO) leads to the activation of microglia featuring a dyshomeostatic transcriptomic profile that resembles disease-associated microglia (DAMs), injury-associated microglia, and aged microglia, suggesting that microglial self-produced TGF-β1 ligands are important in the adult CNS. Interestingly, astrocytes in MG-Tgfb1 iKO mice show a transcriptome profile that closely aligns with A1-like astrocytes. Additionally, using sparse mosaic single-cell microglia iKO of TGF-β1 ligand, we established an autocrine mechanism for TGF-β signaling. Importantly MG-Tgfb1 iKO mice show cognitive deficits, supporting that precise spatial regulation of TGF-β1 ligand derived from microglia is critical for the maintenance of brain homeostasis and normal cognitive function in the adult brain.
Keywords: DAMs; Microglia; TGF-beta; aging; astrocytes; cognitive deficit.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Similar articles
-
Adult microglial TGFβ1 is required for microglia homeostasis via an autocrine mechanism to maintain cognitive function in mice.Nat Commun. 2024 Jun 21;15(1):5306. doi: 10.1038/s41467-024-49596-0. Nat Commun. 2024. PMID: 38906887 Free PMC article.
-
Mouse Bone Marrow-derived Microglia-like Cells Secrete Transforming Growth Factor-β1 and Promote Microglial Aβ Phagocytosis and Reduction of Brain Aβ.Neuroscience. 2020 Jul 1;438:217-228. doi: 10.1016/j.neuroscience.2020.05.004. Neuroscience. 2020. PMID: 32522344
-
Transforming growth factor-β1 acts via TβR-I on microglia to protect against MPP(+)-induced dopaminergic neuronal loss.Brain Behav Immun. 2016 Jan;51:131-143. doi: 10.1016/j.bbi.2015.08.006. Epub 2015 Aug 5. Brain Behav Immun. 2016. PMID: 26254549
-
TGF-β Signaling in Microglia: A Key Regulator of Development, Homeostasis and Reactivity.Biomedicines. 2024 Oct 28;12(11):2468. doi: 10.3390/biomedicines12112468. Biomedicines. 2024. PMID: 39595034 Free PMC article. Review.
-
Astrocytes and the TGF-β1 Pathway in the Healthy and Diseased Brain: a Double-Edged Sword.Mol Neurobiol. 2019 Jul;56(7):4653-4679. doi: 10.1007/s12035-018-1396-y. Epub 2018 Oct 30. Mol Neurobiol. 2019. PMID: 30377983 Review.
References
-
- Paolicelli R. C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011). - PubMed
-
- Nimmerjahn A., Kirchhoff F. & Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). - PubMed
-
- Hu X. et al. Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral ischemia. Stroke 43, 3063–3070 (2012). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
