This is a preprint.
From enhancers to genome conformation: complex transcriptional control underlies expression of a single herpesviral gene
- PMID: 37461644
- PMCID: PMC10350069
- DOI: 10.1101/2023.07.08.548212
From enhancers to genome conformation: complex transcriptional control underlies expression of a single herpesviral gene
Update in
-
Enhancers and genome conformation provide complex transcriptional control of a herpesviral gene.Mol Syst Biol. 2025 Jan;21(1):30-58. doi: 10.1038/s44320-024-00075-0. Epub 2024 Nov 19. Mol Syst Biol. 2025. PMID: 39562742 Free PMC article.
Abstract
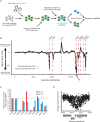
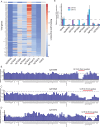
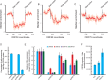
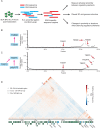
Complex transcriptional control is a conserved feature of both eukaryotes and the viruses that infect them. Here, we illustrate this by combining high-density functional genomics, expression profiling, and viral-specific chromosome conformation capture to define with unprecedented detail the transcriptional regulation of a single gene, ORF68, from Kaposi's sarcoma-associated herpesvirus (KSHV). We first identified seven cis-regulatory regions by densely tiling the ~154 kb KSHV genome with CRISPRi. A parallel Cas9 nuclease screen indicated that three of these regions act as promoters of genes that regulate ORF68. RNA expression profiling demonstrated that three more of these regions act by either repressing or enhancing other distal viral genes involved in ORF68 transcriptional regulation. Finally, we tracked how the 3D structure of the viral genome changes during its lifecycle, revealing that these enhancing regulatory elements are physically closer to their targets when active, and that disrupting some elements caused large-scale changes to the 3D genome. These data enable us to construct a complete model revealing that the mechanistic diversity of this essential regulatory circuit matches that of human genes.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Enhancers and genome conformation provide complex transcriptional control of a herpesviral gene.Mol Syst Biol. 2025 Jan;21(1):30-58. doi: 10.1038/s44320-024-00075-0. Epub 2024 Nov 19. Mol Syst Biol. 2025. PMID: 39562742 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus ORF68 Is a DNA Binding Protein Required for Viral Genome Cleavage and Packaging.J Virol. 2018 Jul 31;92(16):e00840-18. doi: 10.1128/JVI.00840-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875246 Free PMC article.
-
KSHV genome harbors both constitutive and lytically induced enhancers.J Virol. 2024 Jun 13;98(6):e0017924. doi: 10.1128/jvi.00179-24. Epub 2024 May 2. J Virol. 2024. PMID: 38695538 Free PMC article.
-
Next-Generation Sequencing in the Understanding of Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Biology.Viruses. 2016 Mar 31;8(4):92. doi: 10.3390/v8040092. Viruses. 2016. PMID: 27043613 Free PMC article. Review.
-
[Replication Machinery of Kaposi's Sarcoma-associated Herpesvirus and Drug Discovery Research].Yakugaku Zasshi. 2019;139(1):69-73. doi: 10.1248/yakushi.18-00164-2. Yakugaku Zasshi. 2019. PMID: 30606932 Review. Japanese.
References
-
- Qi Y. et al. Latency-associated nuclear antigen inhibits lytic replication of Kaposi’s sarcoma-associated herpesvirus by regulating let-7a/RBPJ signaling. Virology 531, 69–78 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
