This is a preprint.
Critical evaluation of the reliability of DNA methylation probes on the Illumina MethylationEPIC BeadChip microarrays
- PMID: 37461726
- PMCID: PMC10350239
- DOI: 10.21203/rs.3.rs-3068938/v2
Critical evaluation of the reliability of DNA methylation probes on the Illumina MethylationEPIC BeadChip microarrays
Update in
-
Critical evaluation of the reliability of DNA methylation probes on the Illumina MethylationEPIC v1.0 BeadChip microarrays.Epigenetics. 2024 Dec;19(1):2333660. doi: 10.1080/15592294.2024.2333660. Epub 2024 Apr 2. Epigenetics. 2024. PMID: 38564759 Free PMC article.
Abstract
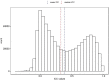
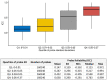
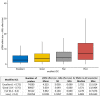
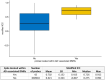
DNA methylation (DNAm) plays a crucial role in a number of complex diseases. However, the reliability of DNAm levels measured using Illumina arrays varies across different probes. Previous research primarily assessed probe reliability by comparing duplicate samples between the 450k-450k or 450k-EPIC platforms, with limited investigations on Illumina EPIC arrays. We conducted a comprehensive assessment of the EPIC array probe reliability using 138 duplicated blood DNAm samples generated by the Alzheimer's Disease Neuroimaging Initiative study. We introduced a novel statistical measure, the modified intraclass correlation, to better account for the disagreement in duplicate measurements. We observed higher reliability in probes with average methylation beta values of 0.2 to 0.8, and lower reliability in type I probes or those within the promoter and CpG island regions. Importantly, we found that probe reliability has significant implications in the analyses of Epigenome-wide Association Studies (EWAS). Higher reliability is associated with more consistent effect sizes in different studies, the identification of differentially methylated regions (DMRs) and methylation quantitative trait locus (mQTLs), and significant correlations with downstream gene expression. Moreover, blood DNAm measurements obtained from probes with higher reliability are more likely to show concordance with brain DNAm measurements. Our findings, which provide crucial reliable information for probes on the EPIC array, will serve as a valuable resource for future DNAm studies.
Keywords: DNA methylation; EPIC array; probe reliability.
Conflict of interest statement
Declaration of Interest Statement The authors report there are no competing interests to declare.
Figures

Similar articles
-
Critical evaluation of the reliability of DNA methylation probes on the Illumina MethylationEPIC v1.0 BeadChip microarrays.Epigenetics. 2024 Dec;19(1):2333660. doi: 10.1080/15592294.2024.2333660. Epub 2024 Apr 2. Epigenetics. 2024. PMID: 38564759 Free PMC article.
-
Systematic evaluation of DNA methylation age estimation with common preprocessing methods and the Infinium MethylationEPIC BeadChip array.Clin Epigenetics. 2018 Oct 16;10(1):123. doi: 10.1186/s13148-018-0556-2. Clin Epigenetics. 2018. PMID: 30326963 Free PMC article.
-
Low reliability of DNA methylation across Illumina Infinium platforms in cord blood: implications for replication studies and meta-analyses of prenatal exposures.Clin Epigenetics. 2022 Jun 28;14(1):80. doi: 10.1186/s13148-022-01299-3. Clin Epigenetics. 2022. PMID: 35765087 Free PMC article.
-
Further Introduction of DNA Methylation (DNAm) Arrays in Regular Diagnostics.Front Genet. 2022 Jul 4;13:831452. doi: 10.3389/fgene.2022.831452. eCollection 2022. Front Genet. 2022. PMID: 35860466 Free PMC article. Review.
-
Improved filtering of DNA methylation microarray data by detection p values and its impact on downstream analyses.Clin Epigenetics. 2019 Jan 24;11(1):15. doi: 10.1186/s13148-019-0615-3. Clin Epigenetics. 2019. PMID: 30678737 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

