Phosphorylation of Schizosaccharomyces pombe Dss1 mediates direct binding to the ubiquitin-ligase Dma1 in vitro
- PMID: 37463013
- PMCID: PMC10443397
- DOI: 10.1002/pro.4733
Phosphorylation of Schizosaccharomyces pombe Dss1 mediates direct binding to the ubiquitin-ligase Dma1 in vitro
Abstract
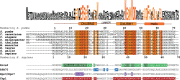
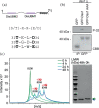
Intrinsically disordered proteins (IDPs) are often multifunctional and frequently posttranslationally modified. Deleted in split hand/split foot 1 (Dss1-Sem1 in budding yeast) is a highly multifunctional IDP associated with a range of protein complexes. However, it remains unknown if the different functions relate to different modified states. In this work, we show that Schizosaccharomyces pombe Dss1 is a substrate for casein kinase 2 in vitro, and we identify three phosphorylated threonines in its linker region separating two known disordered ubiquitin-binding motifs. Phosphorylations of the threonines had no effect on ubiquitin-binding but caused a slight destabilization of the C-terminal α-helix and mediated a direct interaction with the forkhead-associated (FHA) domain of the RING-FHA E3-ubiquitin ligase defective in mitosis 1 (Dma1). The phosphorylation sites are not conserved and are absent in human Dss1. Sequence analyses revealed that the Txx(E/D) motif, which is important for phosphorylation and Dma1 binding, is not linked to certain branches of the evolutionary tree. Instead, we find that the motif appears randomly, supporting the mechanism of ex nihilo evolution of novel motifs. In support of this, other threonine-based motifs, although frequent, are nonconserved in the linker, pointing to additional functions connected to this region. We suggest that Dss1 acts as an adaptor protein that docks to Dma1 via the phosphorylated FHA-binding motifs, while the C-terminal α-helix is free to bind mitotic septins, thereby stabilizing the complex. The presence of Txx(D/E) motifs in the disordered regions of certain septin subunits may be of further relevance to the formation and stabilization of these complexes.
Keywords: FHA; IDP; NMR; Sem1; forkhead-associated domain; phosphorylation; septin; ubiquitin.
© 2023 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures



References
-
- Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, et al. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. - PubMed
-
- Almawi AW, Matthews LA, Guarné A. FHA domains: phosphopeptide binding and beyond. Prog Biophys Mol Biol. 2017;127:105–110. - PubMed
-
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. - PubMed
-
- Blom N, Gammeltoft S, Brunak S. Sequence and structure‐based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

