A hybrid and poly-polish workflow for the complete and accurate assembly of phage genomes: a case study of ten przondoviruses
- PMID: 37463032
- PMCID: PMC10438801
- DOI: 10.1099/mgen.0.001065
A hybrid and poly-polish workflow for the complete and accurate assembly of phage genomes: a case study of ten przondoviruses
Abstract
Bacteriophages (phages) within the genus Przondovirus are T7-like podoviruses belonging to the subfamily Studiervirinae, within the family Autographiviridae, and have a highly conserved genome organisation. The genomes of these phages range from 37 to 42 kb in size, encode 50-60 genes and are characterised by the presence of direct terminal repeats (DTRs) flanking the linear chromosome. These DTRs are often deleted during short-read-only and hybrid assemblies. Moreover, long-read-only assemblies are often littered with sequencing and/or assembly errors and require additional curation. Here, we present the isolation and characterisation of ten novel przondoviruses targeting Klebsiella spp. We describe HYPPA, a HYbrid and Poly-polish Phage Assembly workflow, which utilises long-read assemblies in combination with short-read sequencing to resolve phage DTRs and correcting errors, negating the need for laborious primer walking and Sanger sequencing validation. Our assembly workflow utilised Oxford Nanopore Technologies for long-read sequencing for its accessibility, making it the more relevant long-read sequencing technology at this time, and Illumina DNA Prep for short-read sequencing, representing the most commonly used technologies globally. Our data demonstrate the importance of careful curation of phage assemblies before publication, and prior to using them for comparative genomics.
Keywords: Klebsiella; Przondovirus; assembly; bacteriophage; phage; sequencing.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


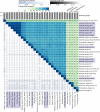
References
-
- ICTV Current ICTV Taxonomy Release. 2022. 2022. [ August 4; 2022 ]. https://ictv.global/taxonomy accessed.
Publication types
MeSH terms
Grants and funding
- BBS/E/F/000PR10349/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 220876/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- BBS/E/F/000PR10356/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/00044409/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10348/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/R015937/1/MRC_/Medical Research Council/United Kingdom
- BB/CCG1860/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 100974 /C/13/Z/WT_/Wellcome Trust/United Kingdom
- BB/R012504/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/T030062/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

