SF3B1 mutation and ATM deletion codrive leukemogenesis via centromeric R-loop dysregulation
- PMID: 37463047
- PMCID: PMC10471171
- DOI: 10.1172/JCI163325
SF3B1 mutation and ATM deletion codrive leukemogenesis via centromeric R-loop dysregulation
Abstract
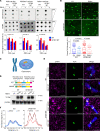
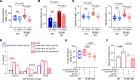
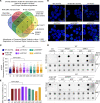
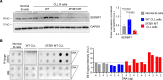
RNA splicing factor SF3B1 is recurrently mutated in various cancers, particularly in hematologic malignancies. We previously reported that coexpression of Sf3b1 mutation and Atm deletion in B cells, but not either lesion alone, leads to the onset of chronic lymphocytic leukemia (CLL) with CLL cells harboring chromosome amplification. However, the exact role of Sf3b1 mutation and Atm deletion in chromosomal instability (CIN) remains unclear. Here, we demonstrated that SF3B1 mutation promotes centromeric R-loop (cen-R-loop) accumulation, leading to increased chromosome oscillation, impaired chromosome segregation, altered spindle architecture, and aneuploidy, which could be alleviated by removal of cen-R-loop and exaggerated by deletion of ATM. Aberrant splicing of key genes involved in R-loop processing underlay augmentation of cen-R-loop, as overexpression of the normal isoform, but not the altered form, mitigated mitotic stress in SF3B1-mutant cells. Our study identifies a critical role of splice variants in linking RNA splicing dysregulation and CIN and highlights cen-R-loop augmentation as a key mechanism for leukemogenesis.
Keywords: Cancer; DNA repair; Oncology.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

