Klebsiella pneumoniae causes bacteremia using factors that mediate tissue-specific fitness and resistance to oxidative stress
- PMID: 37463183
- PMCID: PMC10381055
- DOI: 10.1371/journal.ppat.1011233
Klebsiella pneumoniae causes bacteremia using factors that mediate tissue-specific fitness and resistance to oxidative stress
Abstract
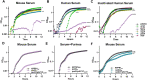
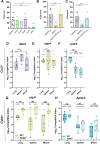
Gram-negative bacteremia is a major cause of global morbidity involving three phases of pathogenesis: initial site infection, dissemination, and survival in the blood and filtering organs. Klebsiella pneumoniae is a leading cause of bacteremia and pneumonia is often the initial infection. In the lung, K. pneumoniae relies on many factors like capsular polysaccharide and branched chain amino acid biosynthesis for virulence and fitness. However, mechanisms directly enabling bloodstream fitness are unclear. Here, we performed transposon insertion sequencing (TnSeq) in a tail-vein injection model of bacteremia and identified 58 K. pneumoniae bloodstream fitness genes. These factors are diverse and represent a variety of cellular processes. In vivo validation revealed tissue-specific mechanisms by which distinct factors support bacteremia. ArnD, involved in Lipid A modification, was required across blood filtering organs and supported resistance to soluble splenic factors. The purine biosynthesis enzyme PurD supported liver fitness in vivo and was required for replication in serum. PdxA, a member of the endogenous vitamin B6 biosynthesis pathway, optimized replication in serum and lung fitness. The stringent response regulator SspA was required for splenic fitness yet was dispensable in the liver. In a bacteremic pneumonia model that incorporates initial site infection and dissemination, splenic fitness defects were enhanced. ArnD, PurD, DsbA, SspA, and PdxA increased fitness across bacteremia phases and each demonstrated unique fitness dynamics within compartments in this model. SspA and PdxA enhanced K. pnuemoniae resistance to oxidative stress. SspA, but not PdxA, specifically resists oxidative stress produced by NADPH oxidase Nox2 in the lung, spleen, and liver, as it was a fitness factor in wild-type but not Nox2-deficient (Cybb-/-) mice. These results identify site-specific fitness factors that act during the progression of Gram-negative bacteremia. Defining K. pneumoniae fitness strategies across bacteremia phases could illuminate therapeutic targets that prevent infection and sepsis.
Copyright: © 2023 Holmes et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have no competing interests to disclose.
Figures




References
-
- WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization; 2017.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

