High-Altitude Andean H194R HIF2A Allele Is a Hypomorphic Allele
- PMID: 37463421
- PMCID: PMC10370452
- DOI: 10.1093/molbev/msad162
High-Altitude Andean H194R HIF2A Allele Is a Hypomorphic Allele
Abstract
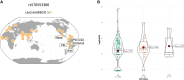
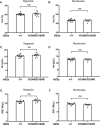
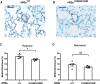
For over 10,000 years, Andeans have resided at high altitude where the partial pressure of oxygen challenges human survival. Recent studies have provided evidence for positive selection acting in Andeans on the HIF2A (also known as EPAS1) locus, which encodes for a central transcription factor of the hypoxia-inducible factor pathway. However, the precise mechanism by which this allele might lead to altitude-adaptive phenotypes, if any, is unknown. By analyzing whole genome sequencing data from 46 high-coverage Peruvian Andean genomes, we confirm evidence for positive selection acting on HIF2A and a unique pattern of variation surrounding the Andean-specific single nucleotide variant (SNV), rs570553380, which encodes for an H194R amino acid substitution in HIF-2α. Genotyping the Andean-associated SNV rs570553380 in a group of 299 Peruvian Andeans from Cerro de Pasco, Peru (4,338 m), reveals a positive association with increased fraction of exhaled nitric oxide, a marker of nitric oxide biosynthesis. In vitro assays show that the H194R mutation impairs binding of HIF-2α to its heterodimeric partner, aryl hydrocarbon receptor nuclear translocator. A knockin mouse model bearing the H194R mutation in the Hif2a gene displays decreased levels of hypoxia-induced pulmonary Endothelin-1 transcripts and protection against hypoxia-induced pulmonary hypertension. We conclude the Andean H194R HIF2A allele is a hypomorphic (partial loss of function) allele.
Keywords: EPAS1; HIF2A; Andean evolution; high-altitude adaptation; hypoxia; hypoxia-inducible factor.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Beall CM. 2014. Adaptation to high altitude: phenotypes and genotypes. Ann Rev Anthropol. 43:251–272.
-
- Beall CM, Laskowski D, Strohl KP, Soria R, Villena M, Vargas E, Alarcon AM, Gonzales C, Erzurum SC. 2001. Pulmonary nitric oxide in mountain dwellers. Nature 414:411–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

