RINT1 deficiency disrupts lipid metabolism and underlies a complex hereditary spastic paraplegia
- PMID: 37463447
- PMCID: PMC10348772
- DOI: 10.1172/JCI162836
RINT1 deficiency disrupts lipid metabolism and underlies a complex hereditary spastic paraplegia
Abstract
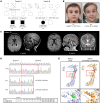
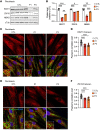
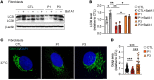
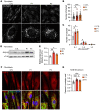
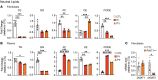
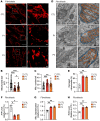
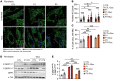
The Rad50 interacting protein 1 (Rint1) is a key player in vesicular trafficking between the ER and Golgi apparatus. Biallelic variants in RINT1 cause infantile-onset episodic acute liver failure (ALF). Here, we describe 3 individuals from 2 unrelated families with novel biallelic RINT1 loss-of-function variants who presented with early onset spastic paraplegia, ataxia, optic nerve hypoplasia, and dysmorphic features, broadening the previously described phenotype. Our functional and lipidomic analyses provided evidence that pathogenic RINT1 variants induce defective lipid-droplet biogenesis and profound lipid abnormalities in fibroblasts and plasma that impact both neutral lipid and phospholipid metabolism, including decreased triglycerides and diglycerides, phosphatidylcholine/phosphatidylserine ratios, and inhibited Lands cycle. Further, RINT1 mutations induced intracellular ROS production and reduced ATP synthesis, affecting mitochondria with membrane depolarization, aberrant cristae ultrastructure, and increased fission. Altogether, our results highlighted the pivotal role of RINT1 in lipid metabolism and mitochondria function, with a profound effect in central nervous system development.
Keywords: Metabolism; Mitochondria; Neurological disorders; Neuroscience.
Conflict of interest statement
Figures