Picolinic acid is a broad-spectrum inhibitor of enveloped virus entry that restricts SARS-CoV-2 and influenza A virus in vivo
- PMID: 37463584
- PMCID: PMC10439173
- DOI: 10.1016/j.xcrm.2023.101127
Picolinic acid is a broad-spectrum inhibitor of enveloped virus entry that restricts SARS-CoV-2 and influenza A virus in vivo
Abstract
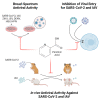
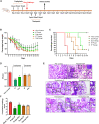
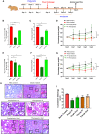
The COVID-19 pandemic highlights an urgent need for effective antivirals. Targeting host processes co-opted by viruses is an attractive antiviral strategy with a high resistance barrier. Picolinic acid (PA) is a tryptophan metabolite endogenously produced in mammals. Here, we report the broad-spectrum antiviral activity of PA against enveloped viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), influenza A virus (IAV), flaviviruses, herpes simplex virus, and parainfluenza virus. Mechanistic studies reveal that PA inhibits enveloped virus entry by compromising viral membrane integrity, inhibiting virus-cellular membrane fusion, and interfering with cellular endocytosis. More importantly, in pre-clinical animal models, PA exhibits promising antiviral efficacy against SARS-CoV-2 and IAV. Overall, our data establish PA as a broad-spectrum antiviral with promising pre-clinical efficacy against pandemic viruses SARS-CoV-2 and IAV.
Keywords: SARS-CoV-2; antiviral; influenza; membrane fusion; picolinic acid; pre-clinical animal models; viral entry.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The Indian Institute of Science has filed patents for the use of PA and its derivatives as broad-spectrum antiviral and listed S.T., R.N., A.B., and R.R. as inventors.
Figures




References
-
- Meganck R.M., Baric R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat. Med. 2021;27:401–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

