Mislocalization of pathogenic RBM20 variants in dilated cardiomyopathy is caused by loss-of-interaction with Transportin-3
- PMID: 37463913
- PMCID: PMC10353998
- DOI: 10.1038/s41467-023-39965-6
Mislocalization of pathogenic RBM20 variants in dilated cardiomyopathy is caused by loss-of-interaction with Transportin-3
Abstract
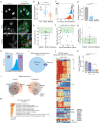
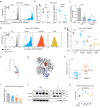
Severe forms of dilated cardiomyopathy (DCM) are associated with point mutations in the alternative splicing regulator RBM20 that are frequently located in the arginine/serine-rich domain (RS-domain). Such mutations can cause defective splicing and cytoplasmic mislocalization, which leads to the formation of detrimental cytoplasmic granules. Successful development of personalized therapies requires identifying the direct mechanisms of pathogenic RBM20 variants. Here, we decipher the molecular mechanism of RBM20 mislocalization and its specific role in DCM pathogenesis. We demonstrate that mislocalized RBM20 RS-domain variants retain their splice regulatory activity, which reveals that aberrant cellular localization is the main driver of their pathological phenotype. A genome-wide CRISPR knockout screen combined with image-enabled cell sorting identified Transportin-3 (TNPO3) as the main nuclear importer of RBM20. We show that the direct RBM20-TNPO3 interaction involves the RS-domain, and is disrupted by pathogenic variants. Relocalization of pathogenic RBM20 variants to the nucleus restores alternative splicing and dissolves cytoplasmic granules in cell culture and animal models. These findings provide proof-of-principle for developing therapeutic strategies to restore RBM20's nuclear localization in RBM20-DCM patients.
© 2023. The Author(s).
Conflict of interest statement
L.M.S. is a co-founder and shareholder of Sophia Genetics. The authors J.K., M.R.M., K.F., M.Go., and L.M.S. filed an invention disclosure describing TNPO3 and restoring nuclear localization of RBM20 variants discussed in this paper (U.S. Provisional Patent Application No. 63/452,252, status: filed: March 15, 2023). M.Go. is an advisor for River Biomedics. A.M.S. received educational grants through his institution from Abbott, Bayer Healthcare, Biosense Webster, Biotronik, Boston Scientific, BMS/Pfizer, and Medtronic; and speaker/advisory board/consulting fees from Bayer Healthcare, Biotronik, Daiichi-Sankyo, Medtronic, Novartis, Pfizer, and Stride Bio Inc. V.N.P. is a consultant and/or advisor for Lexeo Therapeutics, BioMarin, Inc, Viz.ai and Nuevocor. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

