Lampshade web spider Ectatosticta davidi chromosome-level genome assembly provides evidence for its phylogenetic position
- PMID: 37463957
- PMCID: PMC10354039
- DOI: 10.1038/s42003-023-05129-x
Lampshade web spider Ectatosticta davidi chromosome-level genome assembly provides evidence for its phylogenetic position
Abstract
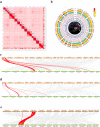
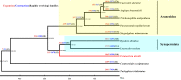
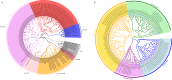
The spider of Ectatosticta davidi, belonging to the lamp-shade web spider family, Hypochilidae, which is closely related to Hypochilidae and Filistatidae and recovered as sister of the rest Araneomorphs spiders. Here we show the final assembled genome of E. davidi with 2.16 Gb in 15 chromosomes. Then we confirm the evolutionary position of Hypochilidae. Moreover, we find that the GMC gene family exhibit high conservation throughout the evolution of true spiders. We also find that the MaSp genes of E. davidi may represent an early stage of MaSp and MiSp genes in other true spiders, while CrSp shares a common origin with AgSp and PySp but differ from MaSp. Altogether, this study contributes to addressing the limited availability of genomic sequences from Hypochilidae spiders, and provides a valuable resource for investigating the genomic evolution of spiders.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
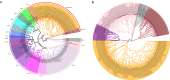


Similar articles
-
Evolution of the karyotype and sex chromosome systems in basal clades of araneomorph spiders (Araneae: Araneomorphae).Chromosome Res. 2006;14(8):859-80. doi: 10.1007/s10577-006-1095-9. Epub 2007 Jan 19. Chromosome Res. 2006. PMID: 17195053
-
Review of the spider genus Ectatosticta Simon, 1892 (Araneae: Hypochilidae) with description of four new species from China.Zootaxa. 2021 Aug 9;5016(4):523-542. doi: 10.11646/zootaxa.5016.4.4. Zootaxa. 2021. PMID: 34810432
-
Mitochondrial phylogenomics provides insights into the phylogeny and evolution of spiders (Arthropoda: Araneae).Zool Res. 2022 Jul 18;43(4):566-584. doi: 10.24272/j.issn.2095-8137.2021.418. Zool Res. 2022. PMID: 35638362 Free PMC article.
-
MASP1 (MBL-associated serine protease 1).Immunobiology. 1998 Aug;199(2):340-7. doi: 10.1016/S0171-2985(98)80038-7. Immunobiology. 1998. PMID: 9777417 Review.
-
Systematics, phylogeny, and evolution of orb-weaving spiders.Annu Rev Entomol. 2014;59:487-512. doi: 10.1146/annurev-ento-011613-162046. Epub 2013 Oct 25. Annu Rev Entomol. 2014. PMID: 24160416 Review.
Cited by
-
Three Novel Spider Genomes Unveil Spidroin Diversification and Hox Cluster Architecture: Ryuthela nishihirai (Liphistiidae), Uloborus plumipes (Uloboridae) and Cheiracanthium punctorium (Cheiracanthiidae).Mol Ecol Resour. 2025 Jan;25(1):e14038. doi: 10.1111/1755-0998.14038. Epub 2024 Oct 22. Mol Ecol Resour. 2025. PMID: 39435585 Free PMC article.
-
Molecular Phylogenetic Relationships Based on Mitogenomes of Spider: Insights Into Evolution and Adaptation to Extreme Environments.Ecol Evol. 2025 Jan 7;15(1):e70774. doi: 10.1002/ece3.70774. eCollection 2025 Jan. Ecol Evol. 2025. PMID: 39781249 Free PMC article.
-
Development and patterning of a highly versatile visual system in spiders.Proc Biol Sci. 2025 Mar;292(2042):20242069. doi: 10.1098/rspb.2024.2069. Epub 2025 Mar 12. Proc Biol Sci. 2025. PMID: 40068820 Free PMC article.
-
Identification and Evolutionary Analysis of the Widely Distributed CAP Superfamily in Spider Venom.Toxins (Basel). 2024 May 24;16(6):240. doi: 10.3390/toxins16060240. Toxins (Basel). 2024. PMID: 38922134 Free PMC article.
-
A trade-off in evolution: the adaptive landscape of spiders without venom glands.Gigascience. 2024 Jan 2;13:giae048. doi: 10.1093/gigascience/giae048. Gigascience. 2024. PMID: 39101784 Free PMC article.
References
-
- WSC. World Spider Catalog. Version 24.0 Natural History Museum Bern. http://wsc.nmbe.ch (2023).
-
- Lehtinen PT. Classification of the cribellate spiders and some allied families, with notes on the evolution of the suborder Araneomorpha. Ann. Zool. Fenn. 1967;4:199–468.
-
- Platnick NI. The hypochiloid spiders: a cladistic analysis, with notes on the Atypoidea (Arachnida, Araneae) Am. Mus. Novit. 1977;2627:1–23.
-
- Coddington JA. Ontogeny and homology in the male palpus of orb-weaving spiders and their relatives, with comments on phylogeny (Araneoclada: Araneoidea, Deinopoidea) Smithson. Contrib. Zool. 1990;496:1–52. doi: 10.5479/si.00810282.496. - DOI
-
- Coddington J. A. Phylogeny and classification of spiders. in Spiders of North America: An identification manual. (eds Ubick, D. et al.) 18–24 (American Arachnological Society, 2005).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

