RUFY3 regulates endolysosomes perinuclear positioning, antigen presentation and migration in activated phagocytes
- PMID: 37463962
- PMCID: PMC10354229
- DOI: 10.1038/s41467-023-40062-x
RUFY3 regulates endolysosomes perinuclear positioning, antigen presentation and migration in activated phagocytes
Abstract
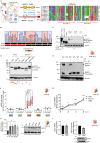
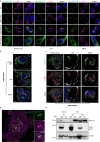
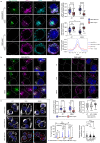
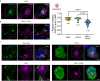
Endo-lysosomes transport along microtubules and clustering in the perinuclear area are two necessary steps for microbes to activate specialized phagocyte functions. We report that RUN and FYVE domain-containing protein 3 (RUFY3) exists as two alternative isoforms distinguishable by the presence of a C-terminal FYVE domain and by their affinity for phosphatidylinositol 3-phosphate on endosomal membranes. The FYVE domain-bearing isoform (iRUFY3) is preferentially expressed in primary immune cells and up-regulated upon activation by microbes and Interferons. iRUFY3 is necessary for ARL8b + /LAMP1+ endo-lysosomes positioning in the pericentriolar organelles cloud of LPS-activated macrophages. We show that iRUFY3 controls macrophages migration, MHC II presentation and responses to Interferon-γ, while being important for intracellular Salmonella replication. Specific inactivation of rufy3 in phagocytes leads to aggravated pathologies in mouse upon LPS injection or bacterial pneumonia. This study highlights the role of iRUFY3 in controlling endo-lysosomal dynamics, which contributes to phagocyte activation and immune response regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

