Disease risk and healthcare utilization among ancestrally diverse groups in the Los Angeles region
- PMID: 37464048
- PMCID: PMC11121511
- DOI: 10.1038/s41591-023-02425-1
Disease risk and healthcare utilization among ancestrally diverse groups in the Los Angeles region
Abstract
An individual's disease risk is affected by the populations that they belong to, due to shared genetics and environmental factors. The study of fine-scale populations in clinical care is important for identifying and reducing health disparities and for developing personalized interventions. To assess patterns of clinical diagnoses and healthcare utilization by fine-scale populations, we leveraged genetic data and electronic medical records from 35,968 patients as part of the UCLA ATLAS Community Health Initiative. We defined clusters of individuals using identity by descent, a form of genetic relatedness that utilizes shared genomic segments arising due to a common ancestor. In total, we identified 376 clusters, including clusters with patients of Afro-Caribbean, Puerto Rican, Lebanese Christian, Iranian Jewish and Gujarati ancestry. Our analysis uncovered 1,218 significant associations between disease diagnoses and clusters and 124 significant associations with specialty visits. We also examined the distribution of pathogenic alleles and found 189 significant alleles at elevated frequency in particular clusters, including many that are not regularly included in population screening efforts. Overall, this work progresses the understanding of health in understudied communities and can provide the foundation for further study into health inequities.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
C.R.G. owns stock in 23andMe, Inc. E.E.K. has received personal fees from Regeneron Pharmaceuticals, 23andMe, Allelica and Illumina; has received research funding from Allelica; and serves on the advisory boards for Encompass Biosciences, Overtone and Galateo Bio. All other authors declare no competing interests.
Figures
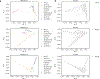










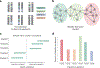

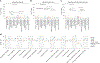
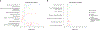

Comment in
-
Leveraging fine-scale population structures for precision healthcare.Nat Med. 2023 Jul;29(7):1611-1612. doi: 10.1038/s41591-023-02423-3. Nat Med. 2023. PMID: 37464034 No abstract available.
References
-
- Fiscella K & Williams DR Health disparities based on socioeconomic inequities: implications for urban health care. Acad. Med. 79, 1139–1147 (2004). - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HL155024/HL/NHLBI NIH HHS/United States
- R01 CA227237/CA/NCI NIH HHS/United States
- R01 GM142112/GM/NIGMS NIH HHS/United States
- F31 NS122538/NS/NINDS NIH HHS/United States
- R01 HG011345/HG/NHGRI NIH HHS/United States
- R01 HL151152/HL/NHLBI NIH HHS/United States
- K01 AG072044/AG/NIA NIH HHS/United States
- U01 HG009080/HG/NHGRI NIH HHS/United States
- R01 MH122688/MH/NIMH NIH HHS/United States
- DP5 OD024579/OD/NIH HHS/United States
- U01 HG011715/HG/NHGRI NIH HHS/United States
- R01 ES029929/ES/NIEHS NIH HHS/United States
- R01 HG006399/HG/NHGRI NIH HHS/United States
- T32 HG002536/HG/NHGRI NIH HHS/United States
- R01 HG010297/HG/NHGRI NIH HHS/United States
- R56 MD013312/MD/NIMHD NIH HHS/United States

