The Role of a Loop in the Non-catalytic Domain B on the Hydrolysis/Transglycosylation Specificity of the 4-α-Glucanotransferase from Thermotoga maritima
- PMID: 37464145
- PMCID: PMC10480278
- DOI: 10.1007/s10930-023-10136-2
The Role of a Loop in the Non-catalytic Domain B on the Hydrolysis/Transglycosylation Specificity of the 4-α-Glucanotransferase from Thermotoga maritima
Abstract
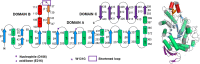
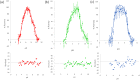
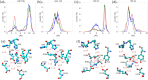
The mechanism by which glycoside hydrolases control the reaction specificity through hydrolysis or transglycosylation is a key element embedded in their chemical structures. The determinants of reaction specificity seem to be complex. We looked for structural differences in domain B between the 4-α-glucanotransferase from Thermotoga maritima (TmGTase) and the α-amylase from Thermotoga petrophila (TpAmylase) and found a longer loop in the former that extends towards the active site carrying a W residue at its tip. Based on these differences we constructed the variants W131G and the partial deletion of the loop at residues 120-124/128-131, which showed a 11.6 and 11.4-fold increased hydrolysis/transglycosylation (H/T) ratio relative to WT protein, respectively. These variants had a reduction in the maximum velocity of the transglycosylation reaction, while their affinity for maltose as the acceptor was not substantially affected. Molecular dynamics simulations allow us to rationalize the increase in H/T ratio in terms of the flexibility near the active site and the conformations of the catalytic acid residues and their associated pKas.
Keywords: Glucanotransferase; Glycosidases; Hydrolysis; Reaction-specificity; Transglycosylation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Crystal structure of Thermotoga maritima 4-alpha-glucanotransferase and its acarbose complex: implications for substrate specificity and catalysis.J Mol Biol. 2002 Aug 2;321(1):149-62. doi: 10.1016/s0022-2836(02)00570-3. J Mol Biol. 2002. PMID: 12139940
-
Modulating Glycoside Hydrolase Activity between Hydrolysis and Transfer Reactions Using an Evolutionary Approach.Molecules. 2021 Oct 30;26(21):6586. doi: 10.3390/molecules26216586. Molecules. 2021. PMID: 34770995 Free PMC article.
-
Purification and characterization of a novel intracellular α-amylase with a wide variety of substrates hydrolysis and transglycosylation activity from Paenibacillus sp. SSG-1.Protein Expr Purif. 2018 Apr;144:62-70. doi: 10.1016/j.pep.2016.04.007. Epub 2016 Apr 20. Protein Expr Purif. 2018. PMID: 27108054
-
Amyrel, a novel glucose-forming α-amylase from Drosophila with 4-α-glucanotransferase activity by disproportionation and hydrolysis of maltooligosaccharides.Glycobiology. 2021 Sep 20;31(9):1134-1144. doi: 10.1093/glycob/cwab036. Glycobiology. 2021. PMID: 33978737
-
Identification of Thermotoga maritima MSB8 GH57 α-amylase AmyC as a glycogen-branching enzyme with high hydrolytic activity.Appl Microbiol Biotechnol. 2019 Aug;103(15):6141-6151. doi: 10.1007/s00253-019-09938-1. Epub 2019 Jun 13. Appl Microbiol Biotechnol. 2019. PMID: 31190240 Free PMC article.
Cited by
-
Structures of α-galactosaminidases from the CAZy GH114 family and homologs defining a new GH191 family of glycosidases.Acta Crystallogr D Struct Biol. 2025 May 1;81(Pt 5):234-251. doi: 10.1107/S2059798325002864. Epub 2025 Apr 15. Acta Crystallogr D Struct Biol. 2025. PMID: 40232846 Free PMC article.
References
-
- Tran LT, Blay V, Luang S, et al. Engineering faster transglycosidases and their acceptor specificity. Green Chem. 2019;21:2823–2836. doi: 10.1039/C9GC00621D. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

