Early application of metagenomics next-generation sequencing may significantly reduce unnecessary consumption of antibiotics in patients with fever of unknown origin
- PMID: 37464295
- PMCID: PMC10354914
- DOI: 10.1186/s12879-023-08417-3
Early application of metagenomics next-generation sequencing may significantly reduce unnecessary consumption of antibiotics in patients with fever of unknown origin
Abstract
Background: Metagenomic next-generation sequencing (mNGS) is a novel nucleic acid method for the detection of unknown and difficult pathogenic microorganisms, and its application in the etiological diagnosis of fever of unknown origin (FUO) is less reported. We aimed to comprehensively assess the value of mNGS in the etiologic diagnosis of FUO by the pathogen spectrum and diagnostic performance, and to investigate whether it is different in the time to diagnosis, length of hospitalization, antibiotic consumption and cost between FUO patients with and without early application of mNGS.
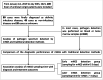
Methods: A total of 149 FUO inpatients underwent both mNGS and routine pathogen detection was retrospectively analyzed. The diagnostic performance of mNGS, culture and CMTs for the final clinical diagnosis was evaluated by using sensitivity, specificity, positive predictive value, negative predictive value and total conforming rate. Patients were furtherly divided into two groups: the earlier mNGS detection group (sampling time: 0 to 3 days of the admission) and the later mNGS detection group (sampling time: after 3 days of the admission). The length of hospital stay, time spent on diagnosis, cost and consumption of antibiotics were compared between the two groups.
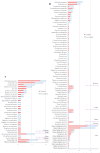
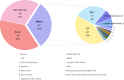
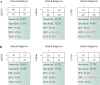
Results: Compared with the conventional microbiological methods, mNGS detected much more species and had the higher negative predictive (67.6%) and total conforming rate (65.1%). Patients with mNGS sampled earlier had a significantly shorter time to diagnosis (6.05+/-6.23 vs. 10.5+/-6.4 days, P < 0.001) and days of hospital stay (13.7+/-20.0 vs. 30.3 +/-26.9, P < 0.001), as well as a significantly less consumption (13.3+/-7.8 vs. 19.5+/-8.0, P < 0.001) and cost (4543+/-7326 vs. 9873 +/- 9958 China Yuan [CNY], P = 0.001) of antibiotics compared with the patients sampled later.
Conclusions: mNGS could significantly improve the detected pathogen spectrum, clinical conforming rate of pathogens while having good negative predictive value for ruling out infections. Early mNGS detection may shorten the diagnosis time and hospitalization days and reduce unnecessary consumption of antibiotics.
Keywords: Consumption of antibiotics; Fever of unknown origin; Metagenomic next-generation sequencing; Pathogen detection; Pathogen spectrum.
© 2023. The Author(s).
Conflict of interest statement
Mingze Tang and Han Xia are employed by Hugobiotech Co., Ltd. All other authors declare no competing interests.
Figures

Similar articles
-
The value of metagenomic next-generation sequencing in the diagnosis of fever of unknown origin.Sci Rep. 2025 Jan 14;15(1):1963. doi: 10.1038/s41598-025-86295-2. Sci Rep. 2025. PMID: 39809928 Free PMC article.
-
Evaluations of Clinical Utilization of Metagenomic Next-Generation Sequencing in Adults With Fever of Unknown Origin.Front Cell Infect Microbiol. 2022 Jan 21;11:745156. doi: 10.3389/fcimb.2021.745156. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35127548 Free PMC article.
-
The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases.BMC Infect Dis. 2021 Jan 13;21(1):62. doi: 10.1186/s12879-020-05746-5. BMC Infect Dis. 2021. PMID: 33435894 Free PMC article.
-
Metagenomic next-generation sequencing in patients with fever of unknown origin: A comprehensive systematic literature review and meta-analysis.Diagn Microbiol Infect Dis. 2024 Oct;110(2):116465. doi: 10.1016/j.diagmicrobio.2024.116465. Epub 2024 Jul 23. Diagn Microbiol Infect Dis. 2024. PMID: 39059148
-
Clinical and diagnostic values of metagenomic next-generation sequencing for infection in hematology patients: a systematic review and meta-analysis.BMC Infect Dis. 2024 Feb 7;24(1):167. doi: 10.1186/s12879-024-09073-x. BMC Infect Dis. 2024. PMID: 38326763 Free PMC article.
Cited by
-
Recommendations for Updating Fever and Inflammation of Unknown Origin From a Modified Delphi Consensus Panel.Open Forum Infect Dis. 2024 Jun 10;11(7):ofae298. doi: 10.1093/ofid/ofae298. eCollection 2024 Jul. Open Forum Infect Dis. 2024. PMID: 38966848 Free PMC article.
-
Update on Fever of Unknown Origin in Children: Focus on Etiologies and Clinical Approach.Children (Basel). 2023 Dec 24;11(1):20. doi: 10.3390/children11010020. Children (Basel). 2023. PMID: 38255334 Free PMC article. Review.
-
Metagenomic next-generation sequencing of plasma cell-free DNA improves the early diagnosis of suspected infections.BMC Infect Dis. 2024 Feb 12;24(1):187. doi: 10.1186/s12879-024-09043-3. BMC Infect Dis. 2024. PMID: 38347444 Free PMC article.
-
The role of metagenomic next-generation sequencing in diagnosing and managing post-kidney transplantation infections.Front Cell Infect Microbiol. 2025 Jan 7;14:1473068. doi: 10.3389/fcimb.2024.1473068. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39839264 Free PMC article. Review.
-
A single-center, retrospective study of hospitalized patients with lower respiratory tract infections: clinical assessment of metagenomic next-generation sequencing and identification of risk factors in patients.Respir Res. 2024 Jun 20;25(1):250. doi: 10.1186/s12931-024-02887-y. Respir Res. 2024. PMID: 38902783 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

