Microbial community structural and functional differentiation in capped thickened oil sands tailings planted with native boreal species
- PMID: 37465026
- PMCID: PMC10350512
- DOI: 10.3389/fmicb.2023.1168653
Microbial community structural and functional differentiation in capped thickened oil sands tailings planted with native boreal species
Abstract
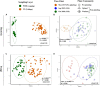
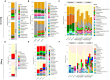
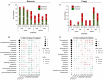
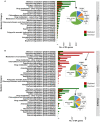
The oil sands mining operations in Alberta have produced billions of m3 of tailings which must be reclaimed and integrated into various mine closure landforms, including terrestrial landforms. Microorganisms play a central role in nutrient cycling during the reclamation of disturbed landscapes, contributing to successful vegetation restoration and long-term sustainability. However, microbial community succession and response in reconstructed and revegetated tailings remain largely unexplored. This study aimed to monitor the structural and functional responses of microbial communities in tailings subjected to different capping and vegetation strategies over two growing seasons (GS). To achieve this, a column-based greenhouse experiment was conducted to investigate microbial communities in tailings that were capped with a layer (10 or 30 cm) of peat-mineral mix (PMM) and planted with either upland or wetland communities. DNA metabarcoding analysis of the bacterial 16S rRNA gene and fungal ITS2 region as well as shotgun metagenomics were used to asses the impact of treatments on microbial taxonomy and functions, respectively. Results showed that tailings microbial diversity and community composition changed considerably after two GS compared to baseline samples, while communities in the PMM capping layer were much more stable. Likewise, several microbial functions were significantly enriched in tailings after two GS. Interestingly, the impact of capping on bacterial communities in tailings varied depending on the plant community, leading to a higher number of differentially abundant taxa and to a decrease in Shannon diversity and evenness in the upland treatment but not in the wetland treatment. Moreover, while capping in the presence of wetland vegetation increased the energy-related metabolic functions (carbon, nitrogen, and sulfur), these functions were depleted by capping in the upland treatment. Fungi represented a small proportion of the microbial community in tailings, but the relative abundance of several taxa changed over time, while the capping treatments favored the growth of some beneficial taxa, notably the root endophyte Serendipita, in both upland and wetland columns. The results suggest that selecting the right combination of capping material and vegetation type may contribute to improve below-ground microbial processes and sustain plant growth in harsh environments such as oil sands tailings.
Keywords: boreal forest; ecological restoration; metabarcoding; metagenomics; oil sands tailings; reclamation; soil microbiology.
Copyright © 2023 Samad, Degenhardt, Séguin, Morency, Gagné and Martineau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alberta Energy Regulator (2020). State of fluid tailings management for mineable oil sands. Calgary, AB: Alberta Energy Regulator.
-
- Alberta Energy Regulator (2022). Directive 085: fluid tailings management for oil sands mining projects. Calgary, AB: Alberta Energy Regulator.
-
- Berta G., Fusconi A., Hooker J. E. (2002). “Arbuscular mycorrhizal modifications to plant root systems: scale, mechanisms and consequences” in Mycorrhizal technology in agriculture: from genes to bioproducts. eds. Gianinazzi S., Schüepp H., Barea J. M., Haselwandter K. (Basel: Birkhäuser; ), 71–85.
-
- Birnbaum C., Bradshaw L. E., Ruthrof K. X., Fontaine J. B. (2017). Topsoil stockpiling in restoration: impact of storage time on plant growth and symbiotic soil biota. Ecol. Res. 35, 237–245. doi: 10.3368/er.35.3.237 - DOI
LinkOut - more resources
Full Text Sources

