Identification of biomarkers associated with diagnosis of postmenopausal osteoporosis patients based on bioinformatics and machine learning
- PMID: 37465165
- PMCID: PMC10352088
- DOI: 10.3389/fgene.2023.1198417
Identification of biomarkers associated with diagnosis of postmenopausal osteoporosis patients based on bioinformatics and machine learning
Abstract
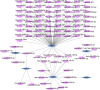
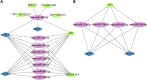
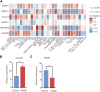
Background: Accumulating evidence suggests that postmenopausal osteoporosis (PMOP) is a common chronic systemic metabolic bone disease, but its specific molecular pathogenesis remains unclear. This study aimed to identify novel genetic diagnostic markers for PMOP. Methods: In this paper, we combined three GEO datasets to identify differentially expressed genes (DEGs) and performed functional enrichment analysis of PMOP-related differential genes. Key genes were analyzed using two machine learning algorithms, namely, LASSO and the Gaussian mixture model, and candidate biomarkers were found after taking the intersection. After further ceRNA network construction, methylation analysis, and immune infiltration analysis, ACACB and WWP1 were finally selected as diagnostic markers. Twenty-four clinical samples were collected, and the expression levels of biomarkers in PMOP were detected by qPCR. Results: We identified 34 differential genes in PMOP. DEG enrichment was mainly related to amino acid synthesis, inflammatory response, and apoptosis. The ceRNA network construction found that XIST-hsa-miR-15a-5p/hsa-miR-15b-5p/hsa-miR-497-5p and hsa-miR-195-5p-WWP1/ACACB may be RNA regulatory pathways regulating PMOP disease progression. ACACB and WWP1 were identified as diagnostic genes for PMOP, and validated in datasets and clinical sample experiments. In addition, these two genes were also significantly associated with immune cells, such as T, B, and NK cells. Conclusion: Overall, we identified two vital diagnostic genes responsible for PMOP. The results may help provide potential immunotherapeutic targets for PMOP.
Keywords: Gene Expression Omnibus; RNA regulatory pathways; diagnosis; machine learning; postmenopausal osteoporosis.
Copyright © 2023 Huang, Ma, Wei, Chen and Chu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
-
Identifying potential ferroptosis key genes for diagnosis and treatment of postmenopausal osteoporosis through competitive endogenous RNA network analysis.Heliyon. 2023 Dec 20;10(1):e23672. doi: 10.1016/j.heliyon.2023.e23672. eCollection 2024 Jan 15. Heliyon. 2023. PMID: 38226266 Free PMC article.
-
Bioinformatics identification and experimental validation of m6A-related diagnostic biomarkers in the subtype classification of blood monocytes from postmenopausal osteoporosis patients.Front Endocrinol (Lausanne). 2023 Mar 8;14:990078. doi: 10.3389/fendo.2023.990078. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36967763 Free PMC article.
-
Olink and gut microbial metabolomics reveal new biomarkers for the prediction and diagnosis of PMOP.J Bone Miner Metab. 2024 Sep;42(5):503-515. doi: 10.1007/s00774-024-01545-z. Epub 2024 Aug 17. J Bone Miner Metab. 2024. PMID: 39153113 Review.
-
HDL-Associated Paraoxonase 1 as a Bridge between Postmenopausal Osteoporosis and Cardiovascular Disease.Chonnam Med J. 2014 Dec;50(3):75-81. doi: 10.4068/cmj.2014.50.3.75. Epub 2014 Dec 17. Chonnam Med J. 2014. PMID: 25568841 Free PMC article. Review.
Cited by
-
The role of WWP1 and WWP2 in bone/cartilage development and diseases.Mol Cell Biochem. 2024 Nov;479(11):2907-2919. doi: 10.1007/s11010-023-04917-7. Epub 2024 Jan 22. Mol Cell Biochem. 2024. PMID: 38252355 Review.
-
To Explore the Key Subgroup and Their Immune Microenvironment During the Formation of Coronary Plaque With scRNA-seq.Cardiol Res Pract. 2025 May 24;2025:3221767. doi: 10.1155/crp/3221767. eCollection 2025. Cardiol Res Pract. 2025. PMID: 40454289 Free PMC article.
-
Heterogeneity of cancer-associated fibroblast subpopulations in prostate cancer: Implications for prognosis and immunotherapy.Transl Oncol. 2025 Feb;52:102255. doi: 10.1016/j.tranon.2024.102255. Epub 2024 Dec 24. Transl Oncol. 2025. PMID: 39721245 Free PMC article.
-
Unveiling the NEFH+ malignant cell subtype: Insights from single-cell RNA sequencing in prostate cancer progression and tumor microenvironment interactions.Front Immunol. 2024 Dec 20;15:1517679. doi: 10.3389/fimmu.2024.1517679. eCollection 2024. Front Immunol. 2024. PMID: 39759507 Free PMC article.
-
Single-cell RNA sequencing revealed PPARG promoted osteosarcoma progression: based on osteoclast proliferation.Front Immunol. 2025 Jan 28;15:1506225. doi: 10.3389/fimmu.2024.1506225. eCollection 2024. Front Immunol. 2025. PMID: 39936154 Free PMC article.
References
LinkOut - more resources
Full Text Sources

