Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field
- PMID: 37465299
- PMCID: PMC10350940
- DOI: 10.1021/acsmedchemlett.3c00104
Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field
Abstract
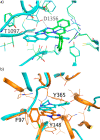
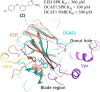
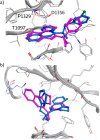
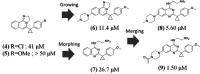
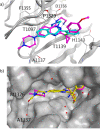
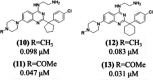
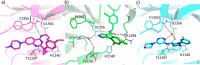
In this study, we describe the rapid identification of potent binders for the WD40 repeat domain (WDR) of DCAF1. This was achieved by two rounds of iterative focused screening of a small set of compounds selected on the basis of internal WDR domain knowledge followed by hit expansion. Subsequent structure-based design led to nanomolar potency binders with a clear exit vector enabling DCAF1-based bifunctional degrader exploration.
© 2023 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
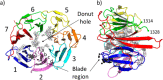

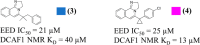
References
-
- Xu C.; Bian C.; Yang W.; Galka M.; Ouyang H.; Chen C.; Qiu W.; Liu H.; Jones A. E.; MacKenzie F.; Pan P.; Li S. S.-C.; Wang H.; Min J. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 19266–19271. 10.1073/pnas.1008937107. - DOI - PMC - PubMed
-
- Tao Y.; Remillard D.; Vinogradova E. V.; Yokoyama M.; Banchenko S.; Schwefel D.; Melillo B.; Schreiber S. L.; Zhang X.; Cravatt B. F. Targeted Protein Degradation by Electrophilic PROTACs that Stereoselectively and Site-Specifically Engage DCAF1. J. Am. Chem. Soc. 2022, 144, 18688–18699. 10.1021/jacs.2c08964. - DOI - PMC - PubMed
-
- Huang Y.; Zhang J.; Yu Z.; Zhang H.; Wang Y.; Lingel A.; Qi W.; Gu J.; Zhao K.; Shultz M. D.; Wang L.; Fu X.; Sun Y.; Zhang Q.; Jiang X.; Zhang J.; Zhang C.; Li L.; Zeng J.; Feng L.; Zhang C.; Liu Y.; Zhang M.; Zhang L.; Zhao M.; Gao Z.; Liu X.; Fang D.; Guo H.; Mi Y.; Gabriel T.; Dillon M. P.; Atadja P.; Oyang C. Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy. J. Med. Chem. 2017, 60, 2215–2226. 10.1021/acs.jmedchem.6b01576. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases

