The Organization and Function of the Phagophore-ER Membrane Contact Sites
- PMID: 37465355
- PMCID: PMC10350784
- DOI: 10.1177/25152564231183898
The Organization and Function of the Phagophore-ER Membrane Contact Sites
Abstract
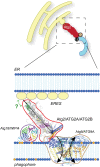
Macroautophagy is characterized by the de novo formation of double-membrane vesicles termed autophagosomes. The precursor structure of autophagosomes is a membrane cistern called phagophore, which elongates through a massive acquisition of lipids until closure. The phagophore establishes membrane-contact sites (MCSs) with the endoplasmic reticulum (ER), where conserved ATG proteins belonging to the ATG9 lipid scramblase, ATG2 lipid transfer and Atg18/WIPI4 β-propeller families concentrate. Several recent in vivo and in vitro studies have uncovered the relevance of these proteins and MCSs in the lipid supply required for autophagosome formation. Although important conceptual advances have been reached, the functional interrelationship between ATG9, ATG2 and Atg18/WIPI4 proteins at the phagophore-ER MCSs and their role in the phagophore expansion are not completely understood. In this review, we describe the current knowledge about the structure, interactions, localizations, and molecular functions of these proteins, with a particular emphasis on the yeast Saccharomyces cerevisiae and mammalian systems.
Keywords: ATG protein; ATG2; ATG9; Atg18/WIPI4; autophagosome; autophagy; lipid transfer; phagophore; scramblase; tethering.
© The Author(s) 2023.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures




References
-
- Bakula D, Muller AJ, Zuleger T, Takacs Z, Franz-Wachtel M, Thost AK, Brigger D, Tschan MP, Frickey T, Robenek H, … (2017). WIPI3 And WIPI4 beta-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nature Communications 8, 15637. doi: 10.1038/ncomms15637 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
