Smooth and accurate predictions of joint contact force time-series in gait using over parameterised deep neural networks
- PMID: 37465692
- PMCID: PMC10350628
- DOI: 10.3389/fbioe.2023.1208711
Smooth and accurate predictions of joint contact force time-series in gait using over parameterised deep neural networks
Abstract
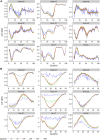
Alterations in joint contact forces (JCFs) are thought to be important mechanisms for the onset and progression of many musculoskeletal and orthopaedic pain disorders. Computational approaches to JCFs assessment represent the only non-invasive means of estimating in-vivo forces; but this cannot be undertaken in free-living environments. Here, we used deep neural networks to train models to predict JCFs, using only joint angles as predictors. Our neural network models were generally able to predict JCFs with errors within published minimal detectable change values. The errors ranged from the lowest value of 0.03 bodyweight (BW) (ankle medial-lateral JCF in walking) to a maximum of 0.65BW (knee VT JCF in running). Interestingly, we also found that over parametrised neural networks by training on longer epochs (>100) resulted in better and smoother waveform predictions. Our methods for predicting JCFs using only joint kinematics hold a lot of promise in allowing clinicians and coaches to continuously monitor tissue loading in free-living environments.
Keywords: deep learning; locomotion; machine learning; musculoskeletal modelling; running biomechanics; walking biomechanics.
Copyright © 2023 Liew, Rügamer, Mei, Altai, Zhu, Zhai and Cortes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Ardestani M. M., Chen Z., Wang L., Lian Q., Liu Y., He J., et al. (2014). Feed forward artificial neural network to predict contact force at medial knee joint: Application to gait modification. Neurocomputing 139, 114–129. 10.1016/j.neucom.2014.02.054 - DOI
-
- Boswell M. A., Uhlrich S. D., Kidziński Ł., Thomas K., Kolesar J. A., Gold G. E., et al. (2021). A neural network to predict the knee adduction moment in patients with osteoarthritis using anatomical landmarks obtainable from 2d video analysis. Osteoarthr. Cartil. 29, 346–356. 10.1016/j.joca.2020.12.017 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

