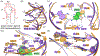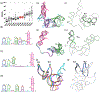RNA target highlights in CASP15: Evaluation of predicted models by structure providers
- PMID: 37466021
- PMCID: PMC10792523
- DOI: 10.1002/prot.26550
RNA target highlights in CASP15: Evaluation of predicted models by structure providers
Abstract
The first RNA category of the Critical Assessment of Techniques for Structure Prediction competition was only made possible because of the scientists who provided experimental structures to challenge the predictors. In this article, these scientists offer a unique and valuable analysis of both the successes and areas for improvement in the predicted models. All 10 RNA-only targets yielded predictions topologically similar to experimentally determined structures. For one target, experimentalists were able to phase their x-ray diffraction data by molecular replacement, showing a potential application of structure predictions for RNA structural biologists. Recommended areas for improvement include: enhancing the accuracy in local interaction predictions and increased consideration of the experimental conditions such as multimerization, structure determination method, and time along folding pathways. The prediction of RNA-protein complexes remains the most significant challenge. Finally, given the intrinsic flexibility of many RNAs, we propose the consideration of ensemble models.
Keywords: CASP; RNA folding; RNA structure prediction; community-wide experiment; cryo-EM; x-ray crystallography.
© 2023 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
Conflict of Interest
All authors declare that they have no competing interests.
Figures