Several Alphaherpesviruses Interact Similarly with the NF-κB Pathway and Suppress NF-κB-Dependent Gene Expression
- PMID: 37466427
- PMCID: PMC10434116
- DOI: 10.1128/spectrum.01421-23
Several Alphaherpesviruses Interact Similarly with the NF-κB Pathway and Suppress NF-κB-Dependent Gene Expression
Abstract
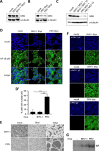
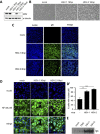
Alphaherpesvirus infection is associated with attenuation of different aspects of the host innate immune response that is elicited to confine primary infections at the mucosal epithelia. Here, we report that infection of epithelial cells with several alphaherpesviruses of different species, including herpes simplex virus 1 and 2 (HSV-1 and HSV-2), feline alphaherpesvirus 1 (FHV-1), and bovine alphaherpesvirus 1 (BoHV-1) results in the inactivation of the responses driven by the nuclear factor kappa B (NF-κB) pathway, considered a pillar of the innate immune response. The mode to interact with and circumvent NF-κB-driven responses in infected epithelial cells is seemingly conserved in human, feline, and porcine alphaherpesviruses, consisting of a persistent activation of the NF-κB cascade but a potent repression of NF-κB-dependent transcription activity, which relies on replication of viral genomes. However, BoHV-1 apparently deviates from the other investigated members of the taxon in this respect, as BoHV-1-infected epithelial cells do not display the persistent NF-κB activation observed for the other alphaherpesviruses. In conclusion, this study suggests that inhibition of NF-κB transcription activity is a strategy used by several alphaherpesviruses to prevent NF-κB-driven responses in infected epithelial cells. IMPORTANCE The current study provides a side-by-side comparison of the interaction of different alphaherpesviruses with NF-κB, a key and central player in the (proinflammatory) innate host response, in infected nontransformed epithelial cell lines. We report that all studied viruses prevent expression of the hallmark NF-κB-dependent gene IκB, often but not always via similar strategies, pointing to suppression of NF-κB-dependent host gene expression in infected epithelial cells as a common and therefore likely important aspect of alphaherpesviruses.
Keywords: NF-κB; alphaherpesvirus; bovine alphaherpesvirus 1; epithelial cells; feline alphaherpesvirus 1; herpes simplex virus; pseudorabies virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Pseudorabies Virus Infection of Epithelial Cells Leads to Persistent but Aberrant Activation of the NF-κB Pathway, Inhibiting Hallmark NF-κB-Induced Proinflammatory Gene Expression.J Virol. 2020 May 4;94(10):e00196-20. doi: 10.1128/JVI.00196-20. Print 2020 May 4. J Virol. 2020. PMID: 32132236 Free PMC article.
-
Pseudorabies Virus Infection Results in a Broad Inhibition of Host Gene Transcription.J Virol. 2022 Jul 13;96(13):e0071422. doi: 10.1128/jvi.00714-22. Epub 2022 Jun 22. J Virol. 2022. PMID: 35730976 Free PMC article.
-
Pseudorabies Virus Infection Triggers NF-κB Activation via the DNA Damage Response but Actively Inhibits NF-κB-Dependent Gene Expression.J Virol. 2021 Nov 23;95(24):e0166621. doi: 10.1128/JVI.01666-21. Epub 2021 Oct 6. J Virol. 2021. PMID: 34613805 Free PMC article.
-
Innate recognition of alphaherpesvirus DNA.Adv Virus Res. 2015;92:63-100. doi: 10.1016/bs.aivir.2014.11.003. Epub 2015 Jan 15. Adv Virus Res. 2015. PMID: 25701886 Review.
-
Alphaherpesvirus in Pets and Livestock.Microorganisms. 2025 Jan 4;13(1):82. doi: 10.3390/microorganisms13010082. Microorganisms. 2025. PMID: 39858850 Free PMC article. Review.
Cited by
-
Skullcapflavone II suppresses TGF-β-induced corneal epithelial mesenchymal transition in vitro.Int J Ophthalmol. 2025 Feb 18;18(2):209-215. doi: 10.18240/ijo.2025.02.02. eCollection 2025. Int J Ophthalmol. 2025. PMID: 39967985 Free PMC article.
-
Apoptosis is mediated by FeHV-1 through the intrinsic pathway and interacts with the autophagic process.Virol J. 2023 Dec 12;20(1):295. doi: 10.1186/s12985-023-02267-w. Virol J. 2023. PMID: 38087282 Free PMC article.
-
Advancements, challenges, and future perspectives in developing feline herpesvirus 1 as a vaccine vector.Front Immunol. 2024 Sep 12;15:1445387. doi: 10.3389/fimmu.2024.1445387. eCollection 2024. Front Immunol. 2024. PMID: 39328406 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

