Open-Source Machine Learning in Computational Chemistry
- PMID: 37466636
- PMCID: PMC10430767
- DOI: 10.1021/acs.jcim.3c00643
Open-Source Machine Learning in Computational Chemistry
Abstract
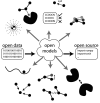
The field of computational chemistry has seen a significant increase in the integration of machine learning concepts and algorithms. In this Perspective, we surveyed 179 open-source software projects, with corresponding peer-reviewed papers published within the last 5 years, to better understand the topics within the field being investigated by machine learning approaches. For each project, we provide a short description, the link to the code, the accompanying license type, and whether the training data and resulting models are made publicly available. Based on those deposited in GitHub repositories, the most popular employed Python libraries are identified. We hope that this survey will serve as a resource to learn about machine learning or specific architectures thereof by identifying accessible codes with accompanying papers on a topic basis. To this end, we also include computational chemistry open-source software for generating training data and fundamental Python libraries for machine learning. Based on our observations and considering the three pillars of collaborative machine learning work, open data, open source (code), and open models, we provide some suggestions to the community.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
ASAS-NANP symposium: mathematical modeling in animal nutrition-Making sense of big data and machine learning: how open-source code can advance training of animal scientists.J Anim Sci. 2023 Jan 3;101:skad317. doi: 10.1093/jas/skad317. J Anim Sci. 2023. PMID: 37997926 Free PMC article.
-
Ichor: A Python library for computational chemistry data management and machine learning force field development.J Comput Chem. 2024 Dec 15;45(32):2912-2928. doi: 10.1002/jcc.27477. Epub 2024 Aug 31. J Comput Chem. 2024. PMID: 39215569
-
PHOTONAI-A Python API for rapid machine learning model development.PLoS One. 2021 Jul 21;16(7):e0254062. doi: 10.1371/journal.pone.0254062. eCollection 2021. PLoS One. 2021. PMID: 34288935 Free PMC article.
-
Deep learning algorithms applied to computational chemistry.Mol Divers. 2024 Aug;28(4):2375-2410. doi: 10.1007/s11030-023-10771-y. Epub 2023 Dec 27. Mol Divers. 2024. PMID: 38151697 Review.
-
Open data and algorithms for open science in AI-driven molecular informatics.Curr Opin Struct Biol. 2023 Apr;79:102542. doi: 10.1016/j.sbi.2023.102542. Epub 2023 Feb 17. Curr Opin Struct Biol. 2023. PMID: 36805192 Review.
Cited by
-
RBPs: an RNA editor's choice.Front Mol Biosci. 2024 Aug 6;11:1454241. doi: 10.3389/fmolb.2024.1454241. eCollection 2024. Front Mol Biosci. 2024. PMID: 39165644 Free PMC article. Review.
-
Data Checking of Asymmetric Catalysis Literature Using a Graph Neural Network Approach.Molecules. 2025 Jan 16;30(2):355. doi: 10.3390/molecules30020355. Molecules. 2025. PMID: 39860224 Free PMC article.
-
Machine Learning Prediction of Quantum Yields and Wavelengths of Aggregation-Induced Emission Molecules.Materials (Basel). 2024 Apr 4;17(7):1664. doi: 10.3390/ma17071664. Materials (Basel). 2024. PMID: 38612177 Free PMC article.
-
The Biology and Biochemistry of Kynurenic Acid, a Potential Nutraceutical with Multiple Biological Effects.Int J Mol Sci. 2024 Aug 21;25(16):9082. doi: 10.3390/ijms25169082. Int J Mol Sci. 2024. PMID: 39201768 Free PMC article. Review.
-
Democratizing cheminformatics: interpretable chemical grouping using an automated KNIME workflow.J Cheminform. 2024 Aug 16;16(1):101. doi: 10.1186/s13321-024-00894-1. J Cheminform. 2024. PMID: 39152469 Free PMC article.
References
-
- Sonnenburg S.; Braun M. L.; Ong C. S.; Bengio S.; Bottou L.; Holmes G.; LeCun Y.; Müller K.-R.; Pereira F.; Rasmussen C. E.; Rätsch G.; Schölkopf B.; Smola A.; Vincent P.; Weston J.; Williamson R. The Need for Open Source Software in Machine Learning. J. Mach. Learn. Res. 2007, 8, 2443–2466.
-
- Langenkamp M.; Yue D. N.. How Open Source Machine Learning Software Shapes AI. In Proceedings of the 2022 AAAI/ACM Conference on AI, Ethics, and Society; ACM: New York, 2022; pp 385–395
-
- Elton D. C.; Boukouvalas Z.; Fuge M. D.; Chung P. W. Deep Learning for Molecular Design – a Review of the State of the Art. Mol. Syst. Des. Eng. 2019, 4, 828–849. 10.1039/C9ME00039A. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

