The pivotal ripening gene SlDML2 participates in regulating disease resistance in tomato
- PMID: 37466912
- PMCID: PMC10579708
- DOI: 10.1111/pbi.14130
The pivotal ripening gene SlDML2 participates in regulating disease resistance in tomato
Abstract
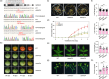
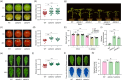
Fruit ripening and disease resistance are two essential biological processes for quality formation and maintenance. DNA methylation, in the form of 5-methylcytosine (5mC), has been elucidated to modulate fruit ripening, but its role in regulating fruit disease resistance remains poorly understood. In this study, we show that mutation of SlDML2, the DNA demethylase gene essential for fruit ripening, affects multiple developmental processes of tomato besides fruit ripening, including seed germination, leaf length and width and flower branching. Intriguingly, loss of SlDML2 function decreased the resistance of tomato fruits against the necrotrophic fungal pathogen Botrytis cinerea. Comparative transcriptomic analysis revealed an obvious transcriptome reprogramming caused by SlDML2 mutation during B. cinerea invasion. Among the thousands of differentially expressed genes, SlβCA3 encoding a β-carbonic anhydrase and SlFAD3 encoding a ω-3 fatty acid desaturase were demonstrated to be transcriptionally activated by SlDML2-mediated DNA demethylation and positively regulate tomato resistance to B. cinerea probably in the same genetic pathway with SlDML2. We further show that the pericarp tissue surrounding B. cinerea infection exhibited a delay in ripening with singnificant decrease in expression of ripening genes that are targeted by SlDML2 and increase in expression of SlβCA3 and SlFAD3. Taken together, our results uncover an essential layer of gene regulation mediated by DNA methylation upon B. cinerea infection and raise the possible that the DNA demethylase gene SlDML2, as a multifunctional gene, participates in modulating the trade-off between fruit ripening and disease resistance.
Keywords: Botrytis cinerea; DNA methylation; SlDML2; defence response; tomato; transcriptome reprogramming.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening.Genome Biol. 2019 Aug 6;20(1):156. doi: 10.1186/s13059-019-1771-7. Genome Biol. 2019. PMID: 31387610 Free PMC article.
-
High mobility group A3 enhances transcription of the DNA demethylase gene SlDML2 to promote tomato fruit ripening.Plant Physiol. 2022 May 3;189(1):315-328. doi: 10.1093/plphys/kiac063. Plant Physiol. 2022. PMID: 35171288 Free PMC article.
-
The role and interaction between transcription factor NAC-NOR and DNA demethylase SlDML2 in the biosynthesis of tomato fruit flavor volatiles.New Phytol. 2022 Sep;235(5):1913-1926. doi: 10.1111/nph.18301. Epub 2022 Jun 30. New Phytol. 2022. PMID: 35686614
-
DNA methylation in tomato fruit ripening.Physiol Plant. 2022 Jan;174(1):e13627. doi: 10.1111/ppl.13627. Physiol Plant. 2022. PMID: 35040145 Review.
-
Transcriptional control of fleshy fruit development and ripening.J Exp Bot. 2014 Aug;65(16):4527-41. doi: 10.1093/jxb/eru316. J Exp Bot. 2014. PMID: 25080453 Review.
Cited by
-
E3 ligase SlCOP1-1 stabilizes transcription factor SlOpaque2 and enhances fruit resistance to Botrytis cinerea in tomato.Plant Physiol. 2024 Oct 1;196(2):1196-1213. doi: 10.1093/plphys/kiae404. Plant Physiol. 2024. PMID: 39077783 Free PMC article.
-
Chemical induction of DNA demethylation by 5-Azacytidine enhances tomato fruit defense against gray mold through dicer-like protein DCL2c.Hortic Res. 2024 Jun 19;11(8):uhae164. doi: 10.1093/hr/uhae164. eCollection 2024 Aug. Hortic Res. 2024. PMID: 39108572 Free PMC article.
-
Unlocking epigenetic breeding potential in tomato and potato.aBIOTECH. 2024 Oct 23;5(4):507-518. doi: 10.1007/s42994-024-00184-2. eCollection 2024 Dec. aBIOTECH. 2024. PMID: 39650134 Free PMC article. Review.
References
-
- Chen, T. , Qin, G. and Tian, S. (2020) Regulatory network of fruit ripening: Current understanding and future challenges. New Phytol. 228, 1219–1226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

