Jurassic NLR: Conserved and dynamic evolutionary features of the atypically ancient immune receptor ZAR1
- PMID: 37467141
- PMCID: PMC10533333
- DOI: 10.1093/plcell/koad175
Jurassic NLR: Conserved and dynamic evolutionary features of the atypically ancient immune receptor ZAR1
Abstract
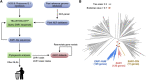
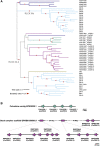
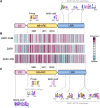
Plant nucleotide-binding leucine-rich repeat (NLR) immune receptors generally exhibit hallmarks of rapid evolution, even at the intraspecific level. We used iterative sequence similarity searches coupled with phylogenetic analyses to reconstruct the evolutionary history of HOPZ-ACTIVATED RESISTANCE1 (ZAR1), an atypically conserved NLR that traces its origin to early flowering plant lineages ∼220 to 150 million yrs ago (Jurassic period). We discovered 120 ZAR1 orthologs in 88 species, including the monocot Colocasia esculenta, the magnoliid Cinnamomum micranthum, and most eudicots, notably the Ranunculales species Aquilegia coerulea, which is outside the core eudicots. Ortholog sequence analyses revealed highly conserved features of ZAR1, including regions for pathogen effector recognition and cell death activation. We functionally reconstructed the cell death activity of ZAR1 and its partner receptor-like cytoplasmic kinase (RLCK) from distantly related plant species, experimentally validating the hypothesis that ZAR1 evolved to partner with RLCKs early in its evolution. In addition, ZAR1 acquired novel molecular features. In cassava (Manihot esculenta) and cotton (Gossypium spp.), ZAR1 carries a C-terminal thioredoxin-like domain, and in several taxa, ZAR1 duplicated into 2 paralog families, which underwent distinct evolutionary paths. ZAR1 stands out among angiosperm NLR genes for having experienced relatively limited duplication and expansion throughout its deep evolutionary history. Nonetheless, ZAR1 also gave rise to noncanonical NLRs with integrated domains and degenerated molecular features.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. S.K. receives funding from industry on NLR biology.
Figures







Comment in
-
Tracing the evolution of the ZAR1 resistosome back to the Jurassic era.Plant Cell. 2023 Sep 27;35(10):3629-3630. doi: 10.1093/plcell/koad193. Plant Cell. 2023. PMID: 37403196 Free PMC article. No abstract available.
References
-
- Adachi H, Sakai T, Kourelis J, Maqbool A, Kamoun S. Jurassic NLR: conserved and dynamic evolutionary features of the atypically ancient immune receptor ZAR1. bioRxiv. 2020. 10.1101/2020.10.12.333484 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

