Transposons are important contributors to gene expression variability under selection in rice populations
- PMID: 37467142
- PMCID: PMC10393045
- DOI: 10.7554/eLife.86324
Transposons are important contributors to gene expression variability under selection in rice populations
Abstract
Transposable elements (TEs) are an important source of genome variability. Here, we analyze their contribution to gene expression variability in rice by performing a TE insertion polymorphism expression quantitative trait locus mapping using expression data from 208 varieties from the Oryza sativa ssp. indica and O. sativa ssp. japonica subspecies. Our data show that TE insertions are associated with changes of expression of many genes known to be targets of rice domestication and breeding. An important fraction of these insertions were already present in the rice wild ancestors, and have been differentially selected in indica and japonica rice populations. Taken together, our results show that small changes of expression in signal transduction genes induced by TE insertions accompany the domestication and adaptation of rice populations.
Keywords: Oriza rufipogon; Oriza sativa; eQTL; evolutionary biology; genetics; genomics; positive selection; transposable elements.
© 2023, Castanera et al.
Conflict of interest statement
RC, NM, SG, MP, JC No competing interests declared
Figures








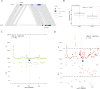

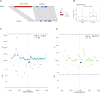

Update of
References
-
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, Levy Y, Harel TH, Shalev-Schlosser G, Amsellem Z, Razifard H, Caicedo AL, Tieman DM, Klee H, Kirsche M, Aganezov S, Ranallo-Benavidez TR, Lemmon ZH, Kim J, Robitaille G, Kramer M, Goodwin S, McCombie WR, Hutton S, Van Eck J, Gillis J, Eshed Y, Sedlazeck FJ, van der Knaap E, Schatz MC, Lippman ZB. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182:145–161. doi: 10.1016/j.cell.2020.05.021. - DOI - PMC - PubMed
-
- Ansari TH, Yamamoto Y, Yoshida T, Miyazaki A, Wang Y. Cultivar differences in the number of differentiated Spikelets and percentage of degenerated Spikelets as determinants of the Spikelet number per Panicle in relation to dry matter production and nitrogen absorption. Soil Science and Plant Nutrition. 2003;49:433–444. doi: 10.1080/00380768.2003.10410029. - DOI
-
- Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, Innan H, Kitano H, Ashikari M, Matsuoka M. Artificial selection for a green revolution gene during Japonica rice Domestication. PNAS. 2011;108:11034–11039. doi: 10.1073/pnas.1019490108. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

