Molecular insights into the inhibitory potential of anthocyanidins on glucokinase regulatory protein
- PMID: 37467274
- PMCID: PMC10355436
- DOI: 10.1371/journal.pone.0288810
Molecular insights into the inhibitory potential of anthocyanidins on glucokinase regulatory protein
Abstract
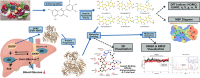
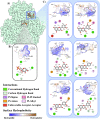
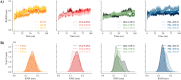
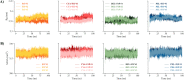
Computational methods were used to investigate six anthocyanidins exhibiting antidiabetic activity by inhibiting glucokinase regulatory protein (GKRP) activity. Density functional theory was used to optimise the geometry of anthocyanidins and calculate their quantum chemical properties. A blind docking method was employed to conduct a molecular docking study, which revealed that delphinidin (Del), cyanidin (Cya), and pelargonidin (Pel) as potential GKRP inhibitors with the lowest binding free energy of -8.7, -8.6, and -8.6 kcal/mol, corresponding to high binding affinity. The molecular dynamics study further verified the blind docking results by showing high GKRP-F1P complex stability and high binding affinity calculated through the MM/GBSA method, upon the binding of pelargonidin. The lower RMSF values of pivotal GK-interacting residues for GKRP-F1P-Pel compared to GKRP-F1P, as a positive control, indicating pelargonidin ability to maintain the inactive conformation of GKRP through the inhibition of GK binding. The key residues that control the binding of the F1P to GKRP and anthocyanidin to GKRP-F1P were also identified in this study. Altogether, pelargonidin is anthocyanidins-derived natural products that have the most potential to act as inhibitors of GKRP and as antidiabetic nutraceuticals.
Copyright: © 2023 Kenneth et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



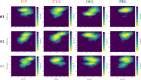
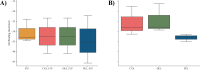
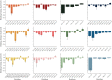
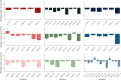
References
-
- Goyal R, Jialal I. Diabetes Mellitus Type 2. StatPearls. Treasure Island: StatPearls Publishing; 2022.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

