Long noncoding RNAs contribute to DNA damage resistance in Arabidopsis thaliana
- PMID: 37467473
- PMCID: PMC10471225
- DOI: 10.1093/genetics/iyad135
Long noncoding RNAs contribute to DNA damage resistance in Arabidopsis thaliana
Abstract
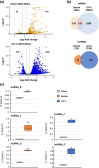
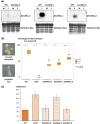
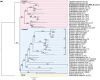
Efficient repair of DNA lesions is essential for the faithful transmission of genetic information between somatic cells and for genome integrity across generations. Plants have multiple, partially redundant, and overlapping DNA repair pathways, probably due to the less constricted germline and the inevitable exposure to light including higher energy wavelengths. Many proteins involved in DNA repair and their mode of actions are well described. In contrast, a role for DNA damage-associated RNA components, evident from many other organisms, is less well understood. Here, we have challenged young Arabidopsis thaliana plants with two different types of genotoxic stress and performed de novo assembly and transcriptome analysis. We identified three long noncoding RNAs (lncRNAs) that are lowly or not expressed under regular conditions but up-regulated or induced by DNA damage. We generated CRISPR/Cas deletion mutants and found that the absence of the lncRNAs impairs the recovery capacity of the plants from genotoxic stress. The genetic loci are highly conserved among world-wide distributed Arabidopsis accessions and within related species in the Brassicaceae group. Together, these results suggest that the lncRNAs have a conserved function in connection with DNA damage and provide a basis for mechanistic analysis of their role.
Keywords: Arabidopsis; Brassicaceae; DNA damage; DNA repair; double strand break; long noncoding RNA; plant.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

