A target discovery pipeline identified ILT3 as a target for immunotherapy of multiple myeloma
- PMID: 37467717
- PMCID: PMC10394163
- DOI: 10.1016/j.xcrm.2023.101110
A target discovery pipeline identified ILT3 as a target for immunotherapy of multiple myeloma
Abstract
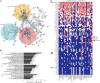
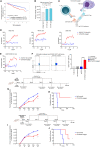
Multiple myeloma (MM) is an incurable malignancy of plasma cells. To identify targets for MM immunotherapy, we develop an integrated pipeline based on mass spectrometry analysis of seven MM cell lines and RNA sequencing (RNA-seq) from 900+ patients. Starting from 4,000+ candidates, we identify the most highly expressed cell surface proteins. We annotate candidate protein expression in many healthy tissues and validate the expression of promising targets in 30+ patient samples with relapsed/refractory MM, as well as in primary healthy hematopoietic stem cells and T cells by flow cytometry. Six candidates (ILT3, SEMA4A, CCR1, LRRC8D, FCRL3, IL12RB1) and B cell maturation antigen (BCMA) present the most favorable profile in malignant and healthy cells. We develop a bispecific T cell engager targeting ILT3 that shows potent killing effects in vitro and decreased tumor burden and prolonged mice survival in vivo, suggesting therapeutic relevance. Our study uncovers MM-associated antigens that hold great promise for immune-based therapies of MM.
Keywords: BCMA; CCR1; FCRL3; IL12RB1; ILT3; Immune-TargetFinder pipeline; LRRC8D; Mass-Spectrometry; SEMA4A; Target discovery strategy; bi-specific T cell engager; immunotherapy; multiple myeloma; primary patient samples; target antigens; validation.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.P. has a patent related to this work: WO2021195536A1 - PCT/US2021/024431.
Figures



References
-
- Friedman K.M., Garrett T.E., Evans J.W., Horton H.M., Latimer H.J., Seidel S.L., Horvath C.J., Morgan R.A. Effective Targeting of Multiple B-Cell Maturation Antigen-Expressing Hematological Malignances by Anti-B-Cell Maturation Antigen Chimeric Antigen Receptor T Cells. Hum. Gene Ther. 2018;29:585–601. doi: 10.1089/hum.2018.001. - DOI - PMC - PubMed
-
- Brudno J.N., Maric I., Hartman S.D., Rose J.J., Wang M., Lam N., Stetler-Stevenson M., Salem D., Yuan C., Pavletic S., et al. T Cells Genetically Modified to Express an Anti-B-Cell Maturation Antigen Chimeric Antigen Receptor Cause Remissions of Poor-Prognosis Relapsed Multiple Myeloma. J. Clin. Oncol. 2018;36:2267–2280. doi: 10.1200/JCO.2018.77.8084. - DOI - PMC - PubMed
-
- Da Vià M.C., Dietrich O., Truger M., Arampatzi P., Duell J., Heidemeier A., Zhou X., Danhof S., Kraus S., Chatterjee M., et al. Homozygous BCMA gene deletion in response to anti-BCMA CAR T cells in a patient with multiple myeloma. Nat. Med. 2021;27:616–619. doi: 10.1038/s41591-021-01245-5. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

