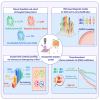
Plasma metabolic fingerprints for large-scale screening and personalized risk stratification of metabolic syndrome
- PMID: 37467725
- PMCID: PMC10394172
- DOI: 10.1016/j.xcrm.2023.101109
Plasma metabolic fingerprints for large-scale screening and personalized risk stratification of metabolic syndrome
Abstract
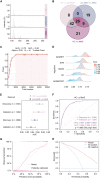
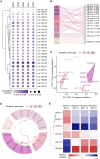
Direct diagnosis and accurate assessment of metabolic syndrome (MetS) allow for prompt clinical interventions. However, traditional diagnostic strategies overlook the complex heterogeneity of MetS. Here, we perform metabolomic analysis in 13,554 participants from the natural cohort and identify 26 hub plasma metabolic fingerprints (PMFs) associated with MetS and its early identification (pre-MetS). By leveraging machine-learning algorithms, we develop robust diagnostic models for pre-MetS and MetS with convincing performance through independent validation. We utilize these PMFs to assess the relative contributions of the four major MetS risk factors in the general population, ranked as follows: hyperglycemia, hypertension, dyslipidemia, and obesity. Furthermore, we devise a personalized three-dimensional plasma metabolic risk (PMR) stratification, revealing three distinct risk patterns. In summary, our study offers effective screening tools for identifying pre-MetS and MetS patients in the general community, while defining the heterogeneous risk stratification of metabolic phenotypes in real-world settings.
Keywords: LDI-MS; machine learning; metabolic syndrome; plasma metabolic fingerprint; risk stratification.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors have filed patents for both the technology and the use of the technology to analyze biofluid samples.
Figures



References
-
- Gurka M.J., Golden S.H., Musani S.K., Sims M., Vishnu A., Guo Y., Cardel M., Pearson T.A., DeBoer M.D. Independent associations between a metabolic syndrome severity score and future diabetes by sex and race: the Atherosclerosis Risk In Communities Study and Jackson Heart Study. Diabetologia. 2017;60:1261–1270. doi: 10.1007/s00125-017-4267-6. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

