Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection
- PMID: 37468466
- PMCID: PMC10356785
- DOI: 10.1038/s41467-023-39859-7
Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection
Abstract
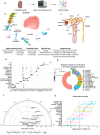
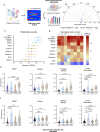
Rejection remains the main cause of premature graft loss after kidney transplantation, despite the use of potent immunosuppression. This highlights the need to better understand the composition and the cell-to-cell interactions of the alloreactive inflammatory infiltrate. Here, we performed droplet-based single-cell RNA sequencing of 35,152 transcriptomes from 16 kidney transplant biopsies with varying phenotypes and severities of rejection and without rejection, and identified cell-type specific gene expression signatures for deconvolution of bulk tissue. A specific association was identified between recipient-derived FCGR3A+ monocytes, FCGR3A+ NK cells and the severity of intragraft inflammation. Activated FCGR3A+ monocytes overexpressed CD47 and LILR genes and increased paracrine signaling pathways promoting T cell infiltration. FCGR3A+ NK cells overexpressed FCRL3, suggesting that antibody-dependent cytotoxicity is a central mechanism of NK-cell mediated graft injury. Multiplexed immunofluorescence using 38 markers on 18 independent biopsy slides confirmed this role of FcγRIII+ NK and FcγRIII+ nonclassical monocytes in antibody-mediated rejection, with specificity to the glomerular area. These results highlight the central involvement of innate immune cells in the pathogenesis of allograft rejection and identify several potential therapeutic targets that might improve allograft longevity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Callemeyn, J. et al. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int.101, 692–710 (2022). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

