A spatially anchored transcriptomic atlas of the human kidney papilla identifies significant immune injury in patients with stone disease
- PMID: 37468493
- PMCID: PMC10356953
- DOI: 10.1038/s41467-023-38975-8
A spatially anchored transcriptomic atlas of the human kidney papilla identifies significant immune injury in patients with stone disease
Abstract
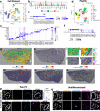
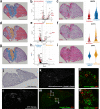
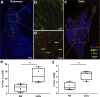
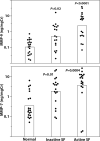
Kidney stone disease causes significant morbidity and increases health care utilization. In this work, we decipher the cellular and molecular niche of the human renal papilla in patients with calcium oxalate (CaOx) stone disease and healthy subjects. In addition to identifying cell types important in papillary physiology, we characterize collecting duct cell subtypes and an undifferentiated epithelial cell type that was more prevalent in stone patients. Despite the focal nature of mineral deposition in nephrolithiasis, we uncover a global injury signature characterized by immune activation, oxidative stress and extracellular matrix remodeling. We also identify the association of MMP7 and MMP9 expression with stone disease and mineral deposition, respectively. MMP7 and MMP9 are significantly increased in the urine of patients with CaOx stone disease, and their levels correlate with disease activity. Our results define the spatial molecular landscape and specific pathways contributing to stone-mediated injury in the human papilla and identify associated urinary biomarkers.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK114923/DK/NIDDK NIH HHS/United States
- UH3 DK114923/DK/NIDDK NIH HHS/United States
- U01 DK133090/DK/NIDDK NIH HHS/United States
- U01 DK114933/DK/NIDDK NIH HHS/United States
- P30 DK079312/DK/NIDDK NIH HHS/United States
- S10 RR023710/RR/NCRR NIH HHS/United States
- U54 DK137328/DK/NIDDK NIH HHS/United States
- R01 DK111651/DK/NIDDK NIH HHS/United States
- U54 DK134301/DK/NIDDK NIH HHS/United States
- P01 DK056788/DK/NIDDK NIH HHS/United States
- R01 DK124776/DK/NIDDK NIH HHS/United States
- U54 HL145608/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

