Influences of chemotype and parental genotype on metabolic fingerprints of tansy plants uncovered by predictive metabolomics
- PMID: 37468576
- PMCID: PMC10356770
- DOI: 10.1038/s41598-023-38790-7
Influences of chemotype and parental genotype on metabolic fingerprints of tansy plants uncovered by predictive metabolomics
Abstract
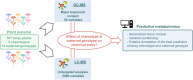
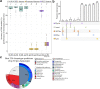
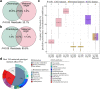
Intraspecific plant chemodiversity shapes plant-environment interactions. Within species, chemotypes can be defined according to variation in dominant specialised metabolites belonging to certain classes. Different ecological functions could be assigned to these distinct chemotypes. However, the roles of other metabolic variation and the parental origin (or genotype) of the chemotypes remain poorly explored. Here, we first compared the capacity of terpenoid profiles and metabolic fingerprints to distinguish five chemotypes of common tansy (Tanacetum vulgare) and depict metabolic differences. Metabolic fingerprints captured higher variation in metabolites while preserving the ability to define chemotypes. These differences might influence plant performance and interactions with the environment. Next, to characterise the influence of the maternal origin on chemodiversity, we performed variation partitioning and generalised linear modelling. Our findings revealed that maternal origin was a higher source of chemical variation than chemotype. Predictive metabolomics unveiled 184 markers predicting maternal origin with 89% accuracy. These markers included, among others, phenolics, whose functions in plant-environment interactions are well established. Hence, these findings place parental genotype at the forefront of intraspecific chemodiversity. We recommend considering this factor when comparing the ecology of various chemotypes. Additionally, the combined inclusion of inherited variation in main terpenoids and other metabolites in computational models may help connect chemodiversity and evolutionary principles.
© 2023. The Author(s).
Conflict of interest statement
We declare no potential conflict of interest. We declare that the experimental research and field studies (including seed collection in Germany) were conducted in accordance with the relevant institutional, national and international guidelines and legislation.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

