A spatially resolved timeline of the human maternal-fetal interface
- PMID: 37468587
- PMCID: PMC10356615
- DOI: 10.1038/s41586-023-06298-9
A spatially resolved timeline of the human maternal-fetal interface
Abstract
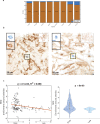
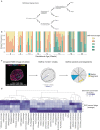
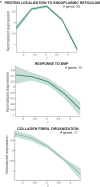
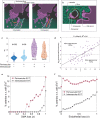
Beginning in the first trimester, fetally derived extravillous trophoblasts (EVTs) invade the uterus and remodel its spiral arteries, transforming them into large, dilated blood vessels. Several mechanisms have been proposed to explain how EVTs coordinate with the maternal decidua to promote a tissue microenvironment conducive to spiral artery remodelling (SAR)1-3. However, it remains a matter of debate regarding which immune and stromal cells participate in these interactions and how this evolves with respect to gestational age. Here we used a multiomics approach, combining the strengths of spatial proteomics and transcriptomics, to construct a spatiotemporal atlas of the human maternal-fetal interface in the first half of pregnancy. We used multiplexed ion beam imaging by time-of-flight and a 37-plex antibody panel to analyse around 500,000 cells and 588 arteries within intact decidua from 66 individuals between 6 and 20 weeks of gestation, integrating this dataset with co-registered transcriptomics profiles. Gestational age substantially influenced the frequency of maternal immune and stromal cells, with tolerogenic subsets expressing CD206, CD163, TIM-3, galectin-9 and IDO-1 becoming increasingly enriched and colocalized at later time points. By contrast, SAR progression preferentially correlated with EVT invasion and was transcriptionally defined by 78 gene ontology pathways exhibiting distinct monotonic and biphasic trends. Last, we developed an integrated model of SAR whereby invasion is accompanied by the upregulation of pro-angiogenic, immunoregulatory EVT programmes that promote interactions with the vascular endothelium while avoiding the activation of maternal immune cells.
© 2023. The Author(s).
Conflict of interest statement
M.A. and is a named inventor on patent US20150287578A1, which covers the mass spectrometry approach utilized by MIBI-TOF to detect elemental reporters in tissue using secondary ion mass spectrometry. M.A. is a board member and shareholder in IonPath, which develops and manufactures the commercial MIBI-TOF platform. All other authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

