Early contact between late farming and pastoralist societies in southeastern Europe
- PMID: 37468624
- PMCID: PMC10412445
- DOI: 10.1038/s41586-023-06334-8
Early contact between late farming and pastoralist societies in southeastern Europe
Abstract
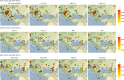
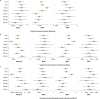
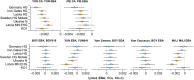
Archaeogenetic studies have described two main genetic turnover events in prehistoric western Eurasia: one associated with the spread of farming and a sedentary lifestyle starting around 7000-6000 BC (refs. 1-3) and a second with the expansion of pastoralist groups from the Eurasian steppes starting around 3300 BC (refs. 4,5). The period between these events saw new economies emerging on the basis of key innovations, including metallurgy, wheel and wagon and horse domestication6-9. However, what happened between the demise of the Copper Age settlements around 4250 BC and the expansion of pastoralists remains poorly understood. To address this question, we analysed genome-wide data from 135 ancient individuals from the contact zone between southeastern Europe and the northwestern Black Sea region spanning this critical time period. While we observe genetic continuity between Neolithic and Copper Age groups from major sites in the same region, from around 4500 BC on, groups from the northwestern Black Sea region carried varying amounts of mixed ancestries derived from Copper Age groups and those from the forest/steppe zones, indicating genetic and cultural contact over a period of around 1,000 years earlier than anticipated. We propose that the transfer of critical innovations between farmers and transitional foragers/herders from different ecogeographic zones during this early contact was integral to the formation, rise and expansion of pastoralist groups around 3300 BC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Similar articles
-
The origins and spread of domestic horses from the Western Eurasian steppes.Nature. 2021 Oct;598(7882):634-640. doi: 10.1038/s41586-021-04018-9. Epub 2021 Oct 20. Nature. 2021. PMID: 34671162 Free PMC article.
-
The genomic history of southeastern Europe.Nature. 2018 Mar 8;555(7695):197-203. doi: 10.1038/nature25778. Epub 2018 Feb 21. Nature. 2018. PMID: 29466330 Free PMC article.
-
Wild cereal grain consumption among Early Holocene foragers of the Balkans predates the arrival of agriculture.Elife. 2021 Dec 1;10:e72976. doi: 10.7554/eLife.72976. Elife. 2021. PMID: 34850680 Free PMC article.
-
The Sequence Analysis of Mitochondrial DNA Revealed Some Major Centers of Horse Domestications: The Archaeologist's Cut.J Equine Vet Sci. 2022 Feb;109:103830. doi: 10.1016/j.jevs.2021.103830. Epub 2021 Dec 4. J Equine Vet Sci. 2022. PMID: 34871751 Review.
-
Where Asia meets Europe - recent insights from ancient human genomics.Ann Hum Biol. 2021 May;48(3):191-202. doi: 10.1080/03014460.2021.1949039. Ann Hum Biol. 2021. PMID: 34459345 Review.
Cited by
-
Cases of trisomy 21 and trisomy 18 among historic and prehistoric individuals discovered from ancient DNA.Nat Commun. 2024 Feb 20;15(1):1294. doi: 10.1038/s41467-024-45438-1. Nat Commun. 2024. PMID: 38378781 Free PMC article.
-
Genetic and geographical origins of Eurasia's influential Yamna culture.Nature. 2025 Mar;639(8053):46-47. doi: 10.1038/d41586-025-00089-0. Nature. 2025. PMID: 39910367 No abstract available.
-
North Pontic crossroads: Mobility in Ukraine from the Bronze Age to the early modern period.Sci Adv. 2025 Jan 10;11(2):eadr0695. doi: 10.1126/sciadv.adr0695. Epub 2025 Jan 8. Sci Adv. 2025. PMID: 39772694 Free PMC article.
-
Demographic interactions between the last hunter-gatherers and the first farmers.Proc Natl Acad Sci U S A. 2025 Apr 8;122(14):e2416221122. doi: 10.1073/pnas.2416221122. Epub 2025 Mar 31. Proc Natl Acad Sci U S A. 2025. PMID: 40163734 Free PMC article.
-
Widespread horse-based mobility arose around 2200 BCE in Eurasia.Nature. 2024 Jul;631(8022):819-825. doi: 10.1038/s41586-024-07597-5. Epub 2024 Jun 6. Nature. 2024. PMID: 38843826 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

