p53 governs an AT1 differentiation programme in lung cancer suppression
- PMID: 37468633
- PMCID: PMC11288504
- DOI: 10.1038/s41586-023-06253-8
p53 governs an AT1 differentiation programme in lung cancer suppression
Abstract
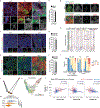
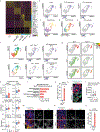
Lung cancer is the leading cause of cancer deaths worldwide1. Mutations in the tumour suppressor gene TP53 occur in 50% of lung adenocarcinomas (LUADs) and are linked to poor prognosis1-4, but how p53 suppresses LUAD development remains enigmatic. We show here that p53 suppresses LUAD by governing cell state, specifically by promoting alveolar type 1 (AT1) differentiation. Using mice that express oncogenic Kras and null, wild-type or hypermorphic Trp53 alleles in alveolar type 2 (AT2) cells, we observed graded effects of p53 on LUAD initiation and progression. RNA sequencing and ATAC sequencing of LUAD cells uncovered a p53-induced AT1 differentiation programme during tumour suppression in vivo through direct DNA binding, chromatin remodelling and induction of genes characteristic of AT1 cells. Single-cell transcriptomics analyses revealed that during LUAD evolution, p53 promotes AT1 differentiation through action in a transitional cell state analogous to a transient intermediary seen during AT2-to-AT1 cell differentiation in alveolar injury repair. Notably, p53 inactivation results in the inappropriate persistence of these transitional cancer cells accompanied by upregulated growth signalling and divergence from lung lineage identity, characteristics associated with LUAD progression. Analysis of Trp53 wild-type and Trp53-null mice showed that p53 also directs alveolar regeneration after injury by regulating AT2 cell self-renewal and promoting transitional cell differentiation into AT1 cells. Collectively, these findings illuminate mechanisms of p53-mediated LUAD suppression, in which p53 governs alveolar differentiation, and suggest that tumour suppression reflects a fundamental role of p53 in orchestrating tissue repair after injury.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures


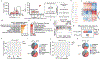











References
-
- Tammemagi MC, McLaughlin JR & Bull SB Meta-analyses of p53 tumor suppressor gene alterations and clinicopathological features in resected lung cancers. Cancer Epidemiol. Biomarkers Prev 8, 625–634 (1999). - PubMed
-
- Jiao XD, Qin BD, You P, Cai J & Zang YS The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 123, 70–75 (2018). - PubMed
-
- Steels E et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur. Respir. J 18, 705–719 (2001). - PubMed
-
- Vaughan AE & Chapman HA Regenerative activity of the lung after epithelial injury. Biochim. Biophys. Acta 1832, 922–930 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

