Comparative evolutionary analyses of eight whitefly Bemisia tabaci sensu lato genomes: cryptic species, agricultural pests and plant-virus vectors
- PMID: 37468834
- PMCID: PMC10357772
- DOI: 10.1186/s12864-023-09474-3
Comparative evolutionary analyses of eight whitefly Bemisia tabaci sensu lato genomes: cryptic species, agricultural pests and plant-virus vectors
Abstract
Background: The group of > 40 cryptic whitefly species called Bemisia tabaci sensu lato are amongst the world's worst agricultural pests and plant-virus vectors. Outbreaks of B. tabaci s.l. and the associated plant-virus diseases continue to contribute to global food insecurity and social instability, particularly in sub-Saharan Africa and Asia. Published B. tabaci s.l. genomes have limited use for studying African cassava B. tabaci SSA1 species, due to the high genetic divergences between them. Genomic annotations presented here were performed using the 'Ensembl gene annotation system', to ensure that comparative analyses and conclusions reflect biological differences, as opposed to arising from different methodologies underpinning transcript model identification.
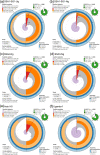
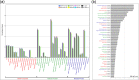
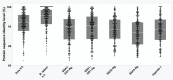
Results: We present here six new B. tabaci s.l. genomes from Africa and Asia, and two re-annotated previously published genomes, to provide evolutionary insights into these globally distributed pests. Genome sizes ranged between 616-658 Mb and exhibited some of the highest coverage of transposable elements reported within Arthropoda. Many fewer total protein coding genes (PCG) were recovered compared to the previously published B. tabaci s.l. genomes and structural annotations generated via the uniform methodology strongly supported a repertoire of between 12.8-13.2 × 103 PCG. An integrative systematics approach incorporating phylogenomic analysis of nuclear and mitochondrial markers supported a monophyletic Aleyrodidae and the basal positioning of B. tabaci Uganda-1 to the sub-Saharan group of species. Reciprocal cross-mating data and the co-cladogenesis pattern of the primary obligate endosymbiont 'Candidatus Portiera aleyrodidarum' from 11 Bemisia genomes further supported the phylogenetic reconstruction to show that African cassava B. tabaci populations consist of just three biological species. We include comparative analyses of gene families related to detoxification, sugar metabolism, vector competency and evaluate the presence and function of horizontally transferred genes, essential for understanding the evolution and unique biology of constituent B. tabaci. s.l species.
Conclusions: These genomic resources have provided new and critical insights into the genetics underlying B. tabaci s.l. biology. They also provide a rich foundation for post-genomic research, including the selection of candidate gene-targets for innovative whitefly and virus-control strategies.
Keywords: Biological species; Cladogenesis; Comparative genomics; Endosymbiont; Genome assembly; Horizontal genes; Phylogenomics; Transposons.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Bellows TS, Jr, Perring TM, Gill RJ, Headrick DH. Description of a species of Bemisia (Homoptera: Aleyrodidae) Ann Entomol Soc Am. 1994;87:195–206. doi: 10.1093/aesa/87.2.195. - DOI
-
- Russell LM. Synonyms of Bemisia tabaci (Gennadius)(Homoptera: Aleyrodidae) Bull Brooklyn Entomol Soc. 1957;52:122–123.
-
- Liu S, Colvin J, De Barro PJ. Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there? J Integr Agric. 2012;11:176–186. doi: 10.1016/S2095-3119(12)60002-1. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

