High-fat diet-disturbed gut microbiota-colonocyte interactions contribute to dysregulating peripheral tryptophan-kynurenine metabolism
- PMID: 37468922
- PMCID: PMC10355067
- DOI: 10.1186/s40168-023-01606-x
High-fat diet-disturbed gut microbiota-colonocyte interactions contribute to dysregulating peripheral tryptophan-kynurenine metabolism
Abstract
Background: Aberrant tryptophan (Trp)-kynurenine (Kyn) metabolism has been implicated in the pathogenesis of human disease. In particular, populations with long-term western-style diets are characterized by an excess of Kyn in the plasma. Host-gut microbiota interactions are dominated by diet and are essential for maintaining host metabolic homeostasis. However, the role of western diet-disturbed gut microbiota-colonocyte interactions in Trp metabolism remains to be elucidated.
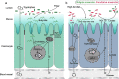
Results: Here, 4-week-old mice were fed with a high-fat diet (HFD), representing a typical western diet, for 4 weeks, and multi-omics approaches were adopted to determine the mechanism by which HFD disrupted gut microbiota-colonocyte interplay causing serum Trp-Kyn metabolism dysfunction. Our results showed that colonocyte-microbiota interactions dominated the peripheral Kyn pathway in HFD mice. Mechanistically, persistent HFD-impaired mitochondrial bioenergetics increased colonic epithelial oxygenation and caused metabolic reprogramming in colonites to support the expansion of Proteobacteria in the colon lumen. Phylum Proteobacteria-derived lipopolysaccharide (LPS) stimulated colonic immune responses to upregulate the indoleamine 2,3-dioxygenase 1 (IDO1)-mediated Kyn pathway, leading to Trp depletion and Kyn accumulation in the circulation, which was further confirmed by transplantation of Escherichia coli (E.coli) indicator strains and colonic IDO1 depletion. Butyrate supplementation promoted mitochondrial functions in colonocytes to remodel the gut microbiota in HFD mice, consequently ameliorating serum Kyn accumulation.
Conclusions: Our results highlighted that HFD disrupted the peripheral Kyn pathway in a gut microbiota-dependent manner and that the continuous homeostasis of gut bacteria-colonocytes interplay played a central role in the regulation of host peripheral Trp metabolism. Meanwhile, this study provided new insights into therapies against western diet-related metabolic disorders. Video Abstract.
Keywords: Gut microbiota; High-fat diet; Kynurenine; Mitochondrial dysfunction; Tryptophan metabolism.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Hezaveh K, Shinde RS, Klötgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. 2022;55(2):324–40.e8. doi: 10.1016/j.immuni.2022.01.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

