PUM1 Promotes Tumor Progression by Activating DEPTOR-Meditated Glycolysis in Gastric Cancer
- PMID: 37469018
- PMCID: PMC10520643
- DOI: 10.1002/advs.202301190
PUM1 Promotes Tumor Progression by Activating DEPTOR-Meditated Glycolysis in Gastric Cancer
Abstract
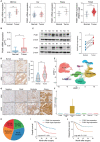
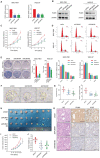
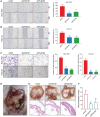
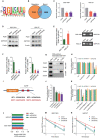
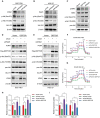
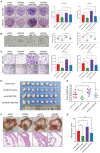
RNA-binding proteins (RBPs) play essential roles in tumorigenesis and progression, but their functions in gastric cancer (GC) remain largely elusive. Here, it is reported that Pumilio 1 (PUM1), an RBP, induces metabolic reprogramming through post-transcriptional regulation of DEP domain-containing mammalian target of rapamycin (mTOR)-interacting protein (DEPTOR) in GC. In clinical samples, elevated expression of PUM1 is associated with recurrence, metastasis, and poor survival. In vitro and in vivo experiments demonstrate that knockdown of PUM1 inhibits the proliferation and metastasis of GC cells. In addition, RNA-sequencing and bioinformatics analyses show that PUM1 is enriched in the glycolysis gene signature. Metabolomics studies confirm that PUM1 deficiency suppresses glycolytic metabolism. Mechanistically, PUM1 binds directly to DEPTOR mRNA pumilio response element to maintain the stability of the transcript and prevent DEPTOR degradation through post-transcriptional pathway. PUM1-mediated DEPTOR upregulation inhibits mTORC1 and alleviates the inhibitory feedback signal transmitted from mTORC1 to PI3K under normal conditions, thus activating the PI3K-Akt signal and glycolysis continuously. Collectively, these results reveal the critical epigenetic role of PUM1 in modulating DEPTOR-dependent GC progression. These conclusions support further clinical investigation of PUM1 inhibitors as a metabolic-targeting treatment strategy for GC.
Keywords: DEPTOR; PI3K-Akt pathway; PUM1; gastric cancer; glycolysis.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest. All these study sponsors have no roles in the study design, in the collection, analysis and interpretation of data.
Figures


References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J. Clin. 2021, 71, 209. - PubMed
-
- Songun I., Putter H., Kranenbarg E. M., Sasako M., van de Velde C. J., Lancet Oncol. 2010, 11, 439. - PubMed
-
- Wilke H., Muro K., Van Cutsem E., Oh S. C., Bodoky G., Shimada Y., Hironaka S., Sugimoto N., Lipatov O., Kim T. Y., Cunningham D., Rougier P., Komatsu Y., Ajani J., Emig M., Carlesi R., Ferry D., Chandrawansa K., Schwartz J. D., Ohtsu A., Group R. S., Lancet Oncol. 2014, 15, 1224. - PubMed
-
- Janjigian Y. Y., Shitara K., Moehler M., Garrido M., Salman P., Shen L., Wyrwicz L., Yamaguchi K., Skoczylas T., Campos Bragagnoli A., Liu T., Schenker M., Yanez P., Tehfe M., Kowalyszyn R., Karamouzis M. V., Bruges R., Zander T., Pazo‐Cid R., Hitre E., Feeney K., Cleary J. M., Poulart V., Cullen D., Lei M., Xiao H., Kondo K., Li M., Ajani J. A., Lancet 2021, 398, 27. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82003104/National Natural Science Foundation of China
- 82073148/National Natural Science Foundation of China
- 81772579/National Natural Science Foundation of China
- 2021B1212040006/Guangdong Provincial Key Laboratory of Digestive Cancer Research
- 2019A1515110632/Guangdong Basic and Applied Basic Research Foundation
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
