Where and when to start: Regulating DNA replication origin activity in eukaryotic genomes
- PMID: 37469113
- PMCID: PMC10361152
- DOI: 10.1080/19491034.2023.2229642
Where and when to start: Regulating DNA replication origin activity in eukaryotic genomes
Abstract
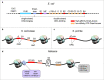
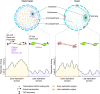
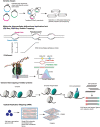
In eukaryotic genomes, hundreds to thousands of potential start sites of DNA replication named origins are dispersed across each of the linear chromosomes. During S-phase, only a subset of origins is selected in a stochastic manner to assemble bidirectional replication forks and initiate DNA synthesis. Despite substantial progress in our understanding of this complex process, a comprehensive 'identity code' that defines origins based on specific nucleotide sequences, DNA structural features, the local chromatin environment, or 3D genome architecture is still missing. In this article, we review the genetic and epigenetic features of replication origins in yeast and metazoan chromosomes and highlight recent insights into how this flexibility in origin usage contributes to nuclear organization, cell growth, differentiation, and genome stability.
Keywords: 3D Genome Organization; DNA replication; chromatin structure; histone modifications; origin clustering; origins of replication; replication timing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
