The effect of the alpha-specific PI3K inhibitor alpelisib combined with anti-HER2 therapy in HER2+/PIK3CA mutant breast cancer
- PMID: 37469415
- PMCID: PMC10353540
- DOI: 10.3389/fonc.2023.1108242
The effect of the alpha-specific PI3K inhibitor alpelisib combined with anti-HER2 therapy in HER2+/PIK3CA mutant breast cancer
Abstract
Background: HER2 is amplified or overexpressed in around 20% of breast cancers (BC). HER2-targeted therapies have significantly improved the prognosis of patients with HER2+ BC, however, de novo and acquired resistance to anti-HER2 treatment is common. Activating mutations in the PIK3CA gene are reported in ∼30% of HER2+ BC and are associated with resistance to anti-HER2 therapies and a poor prognosis. Here, we investigated the in vitro and in vivo antitumor efficacy of the alpha-specific PI3K inhibitor alpelisib alone or in combination with anti-HER2 therapy using a panel of HER2+ BC cell lines. We also generated models of acquired resistance to alpelisib to investigate the mechanisms underlying resistance to alpha-specific PI3K inhibition.
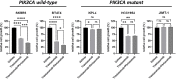
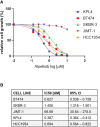
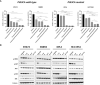
Materials and methods: PIK3CA mutant (HCC1954, KPL4 and JMT1) and wild-type (BT474 and SKBR3) HER2+ BC cell lines were used. The HCC1954 and KPL4 cells were chronically exposed to increasing concentrations of alpelisib or to alpelisib + trastuzumab in order to generate derivatives with acquired resistance to alpelisib (AR) and to alpelisib + trastuzumab (ATR). The transcriptomic profiles of HCC1954, KPL4 and their AR and ATR derivatives were determined by RNA sequencing. Cell growth was assessed by MTT assay. Changes in the protein levels of key PI3K pathway components were assessed by Western blotting. Gene expression, cellular and patients' data from the Cancer Dependency Map (DepMap) and KMPlot datasets were interrogated.
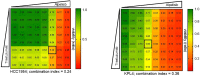
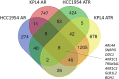
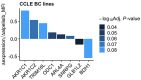
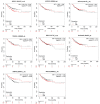
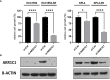
Results: HER2+ BC cell lines harboring activating mutations in PIK3CA were less sensitive to single or dual anti-HER2 blockade compared to PIK3CA wild-type cells. Alpelisib treatment resulted in dose-dependent inhibition of the growth of cells with or without PIK3CA mutations and enhanced the antitumor efficacy of anti-HER2 therapies in vitro. In addition, alpelisib greatly delayed tumor growth of HCC1954 xenografts in vivo. Functional annotation of the significantly differentially expressed genes suggested the common activation of biological processes associated with oxidation reduction, cell proliferation, immune response and RNA synthesis in alpelisib-resistant models compared with native cells. Eight commonly upregulated genes (log2 fold-change >1, False Discovery Rate [FDR] <0.05) in models with acquired resistance to alpelisib or alpelisib + trastuzumab were identified. Among these, AKR1C1 was associated with alpelisib-resistance in vitro and with a poor prognosis in patients with HER2+ BC.
Conclusions: Our findings support the use of an alpha-selective PI3K inhibitor to overcome the therapeutic limitations associated with single or dual HER2 blockade in PIK3CA-mutant HER2+ breast cancer. Future studies are warranted to confirm the potential role of candidate genes/pathways in resistance to alpelisib.
Keywords: AK1RC1; HER2; PIK3CA; alpelisib; breast cancer; resistance.
Copyright © 2023 Cataldo, De Placido, Esposito, Formisano, Arpino, Giuliano, Bianco, De Angelis and Veneziani.
Conflict of interest statement
Author CD reports personal fees from Roche, AstraZeneca, Lilly, GSK, Novartis, Seagen and Pfizer, Advisory Board for Roche, AstraZeneca, Lilly, GSK, Novartis, Seagen and Pfizer, support for attending meetings and/or travel Roche, AstraZeneca, Lilly, GSK, Novartis, Celgene and Pfizer, grants from Novartis. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Xu X, De Angelis C, Burke KA, Nardone A, Hu H, Qin L, et al. HER2 reactivation through acquisition of the HER2 L755S mutation as a mechanism of acquired resistance to HER2-targeted therapy in HER2+ breast cancer. Clin Cancer Res (2017) 23:5123–34. doi: 10.1158/1078-0432.CCR-16-2191 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

