Divide and conquer: genetics, mechanism, and evolution of the ferrous iron transporter Feo in Helicobacter pylori
- PMID: 37469426
- PMCID: PMC10353542
- DOI: 10.3389/fmicb.2023.1219359
Divide and conquer: genetics, mechanism, and evolution of the ferrous iron transporter Feo in Helicobacter pylori
Abstract
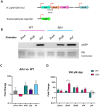
Introduction: Feo is the most widespread and conserved system for ferrous iron uptake in bacteria, and it is important for virulence in several gastrointestinal pathogens. However, its mechanism remains poorly understood. Hitherto, most studies regarding the Feo system were focused on Gammaproteobacterial models, which possess three feo genes (feoA, B, and C) clustered in an operon. We found that the human pathogen Helicobacter pylori possesses a unique arrangement of the feo genes, in which only feoA and feoB are present and encoded in distant loci. In this study, we examined the functional significance of this arrangement.
Methods: Requirement and regulation of the individual H. pylori feo genes were assessed through in vivo assays and gene expression profiling. The evolutionary history of feo was inferred via phylogenetic reconstruction, and AlphaFold was used for predicting the FeoA-FeoB interaction.
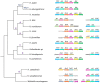
Results and discussion: Both feoA and feoB are required for Feo function, and feoB is likely subjected to tight regulation in response to iron and nickel by Fur and NikR, respectively. Also, we established that feoA is encoded in an operon that emerged in the common ancestor of most, but not all, helicobacters, and this resulted in feoA transcription being controlled by two independent promoters. The H. pylori Feo system offers a new model to understand ferrous iron transport in bacterial pathogens.
Keywords: Feo; Fur; Helicobacter pylori; NikR; Vibrio cholerae; iron transport; nickel; operon.
Copyright © 2023 Gómez-Garzón and Payne.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








Similar articles
-
Disentangling the Evolutionary History of Feo, the Major Ferrous Iron Transport System in Bacteria.mBio. 2022 Feb 22;13(1):e0351221. doi: 10.1128/mbio.03512-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012344 Free PMC article.
-
Vibrio cholerae FeoA, FeoB, and FeoC Interact To Form a Complex.J Bacteriol. 2016 Feb 1;198(7):1160-70. doi: 10.1128/JB.00930-15. J Bacteriol. 2016. PMID: 26833408 Free PMC article.
-
FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB.J Bacteriol. 2013 Nov;195(21):4826-35. doi: 10.1128/JB.00738-13. Epub 2013 Aug 16. J Bacteriol. 2013. PMID: 23955009 Free PMC article.
-
Toward a mechanistic understanding of Feo-mediated ferrous iron uptake.Metallomics. 2018 Jul 18;10(7):887-898. doi: 10.1039/c8mt00097b. Metallomics. 2018. PMID: 29953152 Free PMC article. Review.
-
Bacterial ferrous iron transport: the Feo system.FEMS Microbiol Rev. 2016 Mar;40(2):273-98. doi: 10.1093/femsre/fuv049. Epub 2015 Dec 17. FEMS Microbiol Rev. 2016. PMID: 26684538 Review.
Cited by
-
Challenges and Prospects for Eradication of Helicobacter pylori: Targeting Virulence Factors, Metabolism, and Vaccine Innovation.Pathogens. 2025 Jun 21;14(7):619. doi: 10.3390/pathogens14070619. Pathogens. 2025. PMID: 40732667 Free PMC article. Review.
-
Single-base resolution quantitative genome methylation analysis in the model bacterium Helicobacter pylori by enzymatic methyl sequencing (EM-Seq) reveals influence of strain, growth phase, and methyl homeostasis.BMC Biol. 2024 May 29;22(1):125. doi: 10.1186/s12915-024-01921-1. BMC Biol. 2024. PMID: 38807090 Free PMC article.
References
-
- Agriesti F., Roncarati D., Musiani F., Del Campo C., Iurlaro M., Sparla F., et al. (2014). FeON-FeOFF: the Helicobacter pylori Fur regulator commutates iron-responsive transcription by discriminative readout of opposed DNA grooves. Nucleic Acids Res. 42, 3138–3151. doi: 10.1093/nar/gkt1258, PMID: - DOI - PMC - PubMed
-
- Bereswill S., Greiner S., Van Vliet A. H. M., Waidner B., Fassbinder F., Schiltz E., et al. (2000). Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182, 5948–5953. doi: 10.1128/JB.182.21.5948-5953.2000, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

